| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
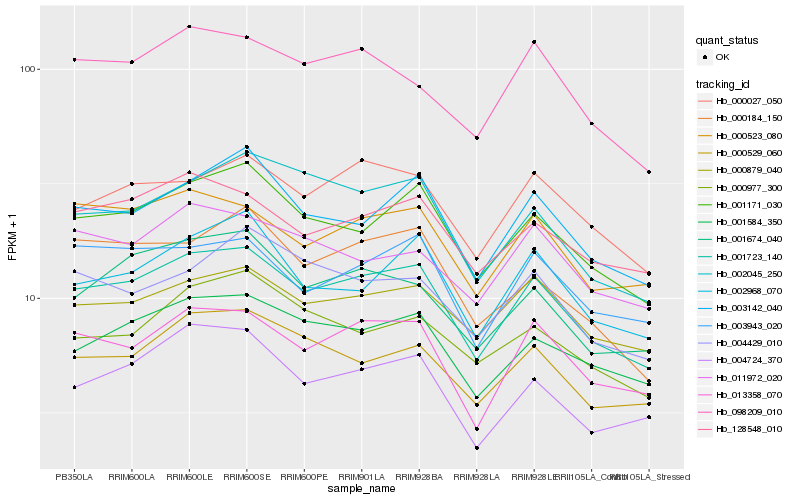
Hb_001723_140 |
0.0 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 2 |
Hb_001171_030 |
0.0497508664 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 3 |
Hb_013358_070 |
0.0521294957 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_001584_350 |
0.0566235143 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 5 |
Hb_003142_040 |
0.059541086 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 6 |
Hb_000027_050 |
0.0624918338 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000529_060 |
0.063320678 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000523_080 |
0.0636529469 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 9 |
Hb_128548_010 |
0.06435936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000879_040 |
0.0661262428 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 11 |
Hb_001674_040 |
0.0676210236 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 12 |
Hb_003943_020 |
0.0679133259 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002045_250 |
0.0680500768 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 14 |
Hb_004724_370 |
0.0681333845 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 15 |
Hb_002968_070 |
0.0689534332 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 16 |
Hb_098209_010 |
0.0691000121 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 17 |
Hb_000184_150 |
0.0745189309 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 18 |
Hb_000977_300 |
0.0753281502 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 19 |
Hb_011972_020 |
0.0761448527 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_004429_010 |
0.076281868 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |