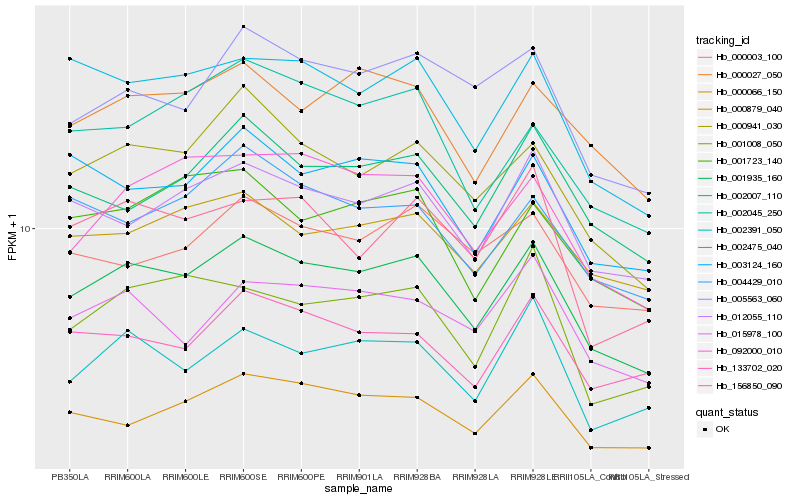
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001935_160 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 2 |
Hb_000941_030 |
0.0643922007 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 3 |
Hb_012055_110 |
0.0681093356 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 4 |
Hb_002475_040 |
0.0706273807 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000027_050 |
0.0727902397 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000066_150 |
0.0747784261 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 7 |
Hb_000879_040 |
0.0749660957 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 8 |
Hb_002007_110 |
0.0766989242 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 9 |
Hb_092000_010 |
0.0767396639 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 10 |
Hb_003124_160 |
0.0767809424 |
- |
- |
dynamin, putative [Ricinus communis] |
| 11 |
Hb_005563_060 |
0.0769626791 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 12 |
Hb_015978_100 |
0.0784613476 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 13 |
Hb_001723_140 |
0.078783956 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_133702_020 |
0.0795297124 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_004429_010 |
0.0798338091 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_002391_050 |
0.0813179595 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 17 |
Hb_001008_050 |
0.0817835348 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 18 |
Hb_000003_100 |
0.0821102057 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 19 |
Hb_002045_250 |
0.0827115234 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 20 |
Hb_156850_090 |
0.0830236354 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |