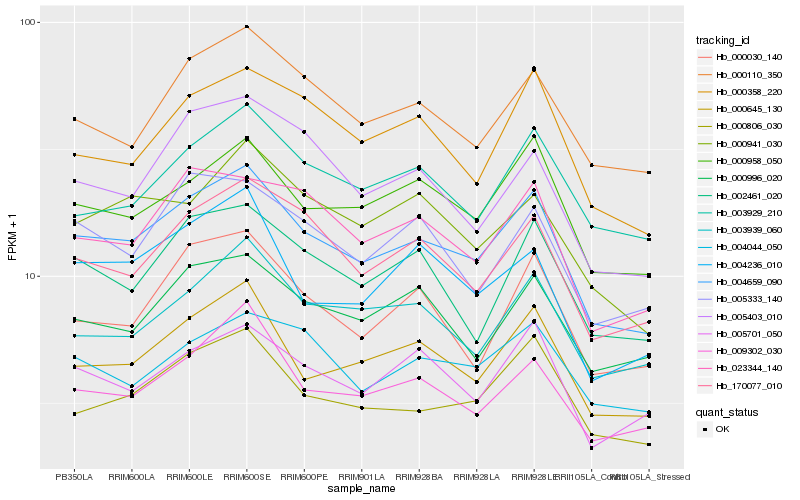
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004659_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 2 |
Hb_000645_130 |
0.0617450527 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_170077_010 |
0.0659044319 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000958_050 |
0.0660336353 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 5 |
Hb_000806_030 |
0.0665003812 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 6 |
Hb_009302_030 |
0.0700147786 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 7 |
Hb_000110_350 |
0.072576488 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_023344_140 |
0.0728032505 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 9 |
Hb_002461_020 |
0.074090909 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000358_220 |
0.0751066385 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 11 |
Hb_005701_050 |
0.0752391296 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 12 |
Hb_003939_060 |
0.0768811511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000996_020 |
0.0777473459 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 14 |
Hb_005333_140 |
0.0784777734 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000941_030 |
0.0785325041 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 16 |
Hb_000030_140 |
0.0788067043 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004236_010 |
0.0797881424 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 18 |
Hb_005403_010 |
0.0798103796 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 19 |
Hb_003929_210 |
0.079899871 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 20 |
Hb_004044_050 |
0.080248376 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |