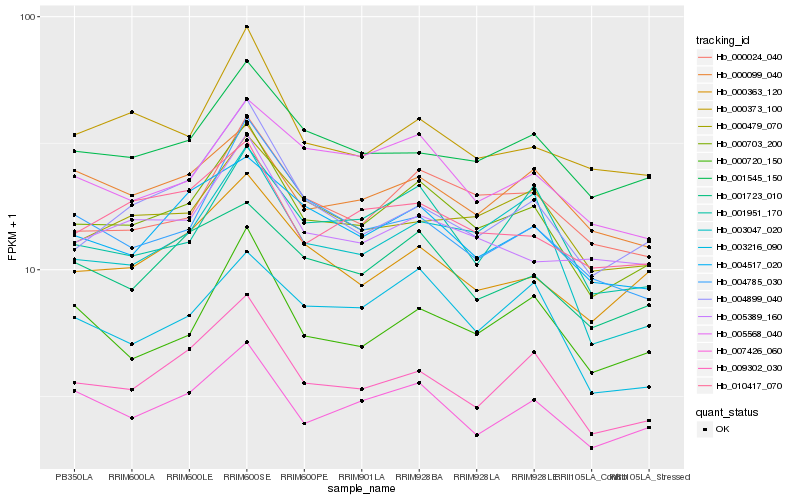
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_200 |
0.0 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_009302_030 |
0.0643202705 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 3 |
Hb_000024_040 |
0.0659126589 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 4 |
Hb_007426_060 |
0.0671656796 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_001545_150 |
0.0708480258 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 6 |
Hb_001723_010 |
0.0726387075 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6 [Jatropha curcas] |
| 7 |
Hb_004517_020 |
0.073342524 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000363_120 |
0.0741924943 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 9 |
Hb_004899_040 |
0.0756898572 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 10 |
Hb_001951_170 |
0.0758257783 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_000479_070 |
0.075885327 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 12 |
Hb_010417_070 |
0.0772915709 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003047_020 |
0.0778072297 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_005568_040 |
0.0779524402 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000373_100 |
0.0779705888 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |
| 16 |
Hb_000720_150 |
0.0785428724 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |
| 17 |
Hb_005389_160 |
0.0795032533 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 18 |
Hb_003216_090 |
0.0804444173 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000099_040 |
0.0811057731 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_004785_030 |
0.0813122819 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1 [Jatropha curcas] |