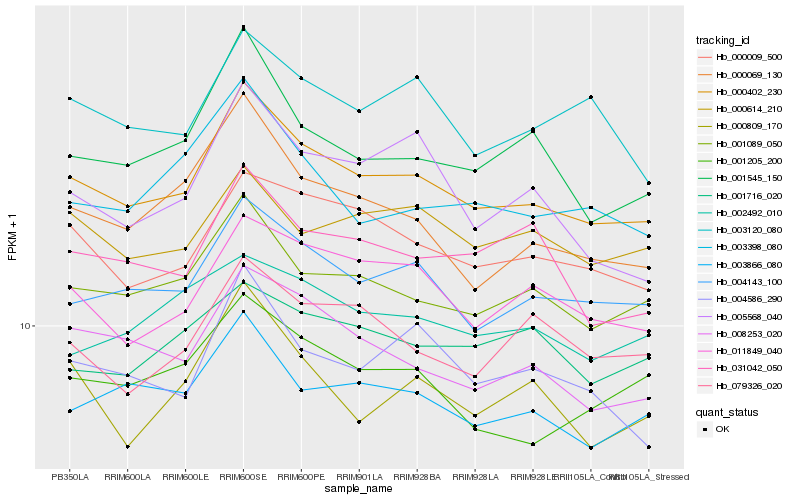
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_230 |
0.0 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004143_100 |
0.0427077122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001089_050 |
0.0442840039 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000009_500 |
0.0497625009 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 5 |
Hb_008253_020 |
0.0526331699 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 6 |
Hb_003866_080 |
0.0530721459 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 7 |
Hb_011849_040 |
0.053190551 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_001545_150 |
0.0560580485 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 9 |
Hb_000069_130 |
0.0602169074 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003120_080 |
0.0621614135 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004586_290 |
0.0625570448 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 12 |
Hb_031042_050 |
0.0627088051 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 13 |
Hb_000614_210 |
0.0628736575 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001205_200 |
0.0630025751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003398_080 |
0.0632230911 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_079326_020 |
0.0651104951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002492_010 |
0.0656241699 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 18 |
Hb_000809_170 |
0.0662216948 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_001716_020 |
0.0677864491 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 20 |
Hb_005568_040 |
0.0693228654 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |