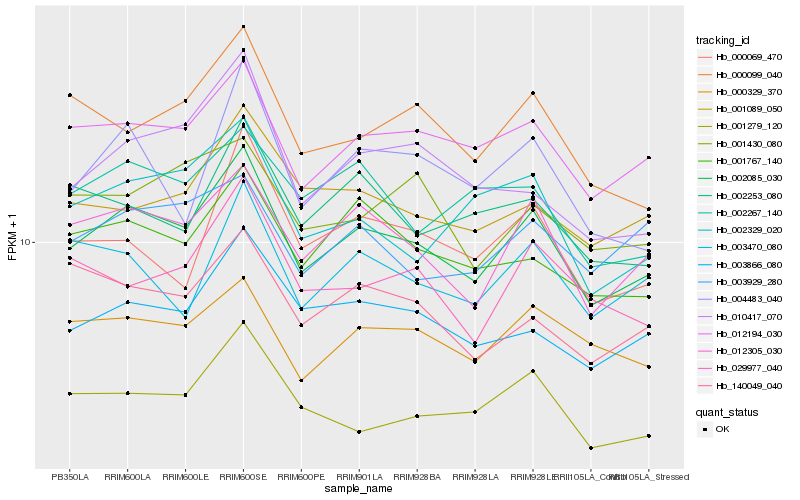
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002085_030 |
0.0 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012305_030 |
0.0463695946 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 3 |
Hb_012194_030 |
0.0485850195 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003866_080 |
0.0580995289 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 5 |
Hb_001767_140 |
0.0605233672 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 6 |
Hb_001279_120 |
0.0607389968 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 7 |
Hb_000329_370 |
0.0631903351 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 8 |
Hb_140049_040 |
0.0667400787 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 9 |
Hb_001089_050 |
0.0668825825 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_000099_040 |
0.0670933768 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_029977_040 |
0.067986134 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 12 |
Hb_003470_080 |
0.0687871915 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002267_140 |
0.069535273 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 14 |
Hb_002329_020 |
0.0696688637 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 15 |
Hb_010417_070 |
0.069887429 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004483_040 |
0.0701063814 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 17 |
Hb_003929_280 |
0.0704097822 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 18 |
Hb_002253_080 |
0.0705834266 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 19 |
Hb_000069_470 |
0.0706325526 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001430_080 |
0.0710242908 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |