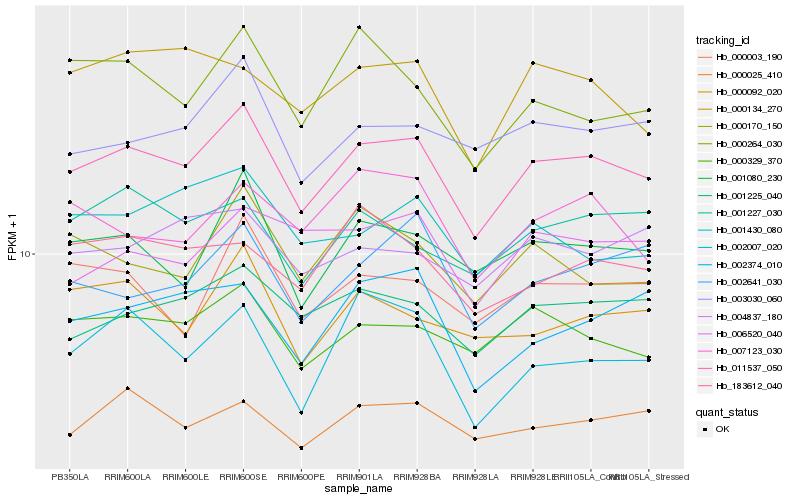
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011537_050 |
0.0 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000329_370 |
0.0628960054 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 3 |
Hb_001225_040 |
0.0640729014 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 4 |
Hb_001080_230 |
0.0652883738 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 5 |
Hb_003030_060 |
0.0702787513 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 6 |
Hb_000025_410 |
0.0709104411 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 7 |
Hb_000003_190 |
0.0731658495 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_001227_030 |
0.0755424906 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000170_150 |
0.0765389405 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000092_020 |
0.0770888574 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_002374_010 |
0.077134949 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001430_080 |
0.0773536741 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 13 |
Hb_004837_180 |
0.0777721087 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000264_030 |
0.077807405 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 15 |
Hb_002641_030 |
0.0787971114 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 16 |
Hb_000134_270 |
0.078954472 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 17 |
Hb_002007_020 |
0.0795241883 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like SIM [Jatropha curcas] |
| 18 |
Hb_183612_040 |
0.0796302446 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 19 |
Hb_007123_030 |
0.0798405991 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 20 |
Hb_006520_040 |
0.0803961938 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |