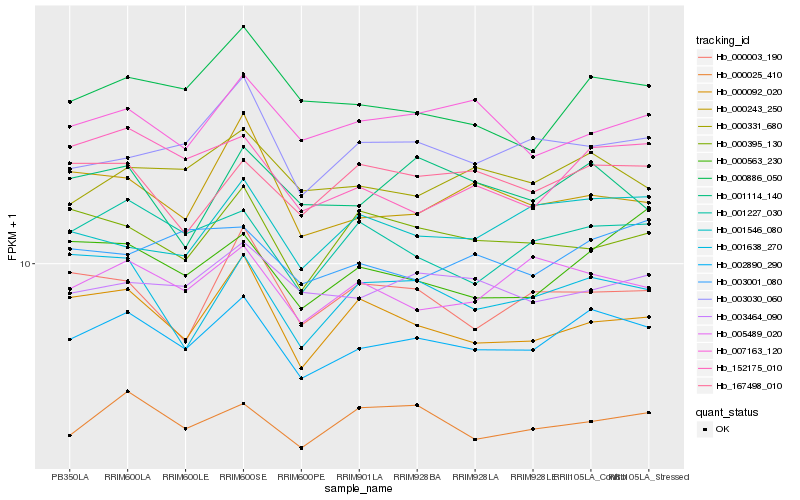
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_290 |
0.0 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001227_030 |
0.0631726434 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005489_020 |
0.0643647168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000331_680 |
0.0652068734 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 5 |
Hb_000025_410 |
0.0664715677 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |
| 6 |
Hb_152175_010 |
0.0665958275 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 7 |
Hb_003464_090 |
0.0693024081 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 8 |
Hb_167498_010 |
0.0700844774 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 9 |
Hb_000886_050 |
0.0704066928 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 10 |
Hb_000092_020 |
0.0714611859 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_007163_120 |
0.073983528 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 12 |
Hb_003030_060 |
0.0741128868 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 13 |
Hb_003001_080 |
0.0745556738 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 14 |
Hb_001546_080 |
0.0755356437 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 15 |
Hb_001638_270 |
0.0756009025 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 8 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_001114_140 |
0.0758170525 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 17 |
Hb_000563_230 |
0.0765071455 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 18 |
Hb_000395_130 |
0.0776860454 |
- |
- |
PREDICTED: N6-adenosine-methyltransferase MT-A70-like [Jatropha curcas] |
| 19 |
Hb_000243_250 |
0.0783774641 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 20 |
Hb_000003_190 |
0.07852254 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |