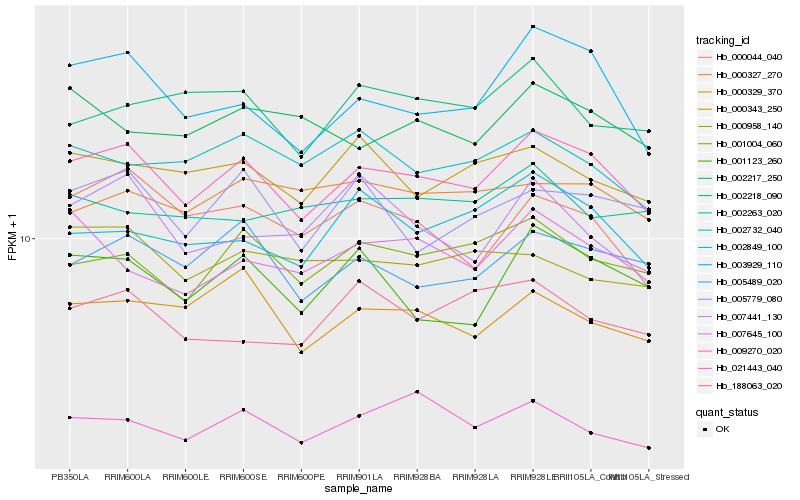
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021443_040 |
0.0 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 2 |
Hb_003929_110 |
0.0558189237 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_005489_020 |
0.0620003068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001004_060 |
0.0624141111 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 5 |
Hb_001123_260 |
0.0689458661 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 6 |
Hb_000329_370 |
0.0695601753 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 7 |
Hb_000343_250 |
0.070995818 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 8 |
Hb_005779_080 |
0.0717594079 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009270_020 |
0.0720845194 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 10 |
Hb_000044_040 |
0.0721434141 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 11 |
Hb_002849_100 |
0.0727532338 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002732_040 |
0.0748711856 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007645_100 |
0.0763378692 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002218_090 |
0.0776129948 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000327_270 |
0.0777320351 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_002217_250 |
0.0782968573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_188063_020 |
0.0787827363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000958_140 |
0.0815127108 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |
| 19 |
Hb_007441_130 |
0.0831637634 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_002263_020 |
0.0839581889 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |