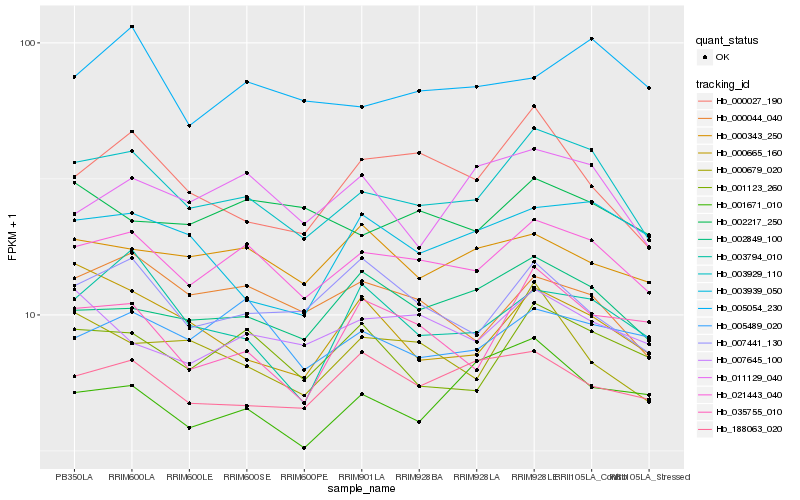
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003929_110 |
0.0 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_021443_040 |
0.0558189237 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 3 |
Hb_001123_260 |
0.0866652846 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 4 |
Hb_002849_100 |
0.0886439171 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000679_020 |
0.0930632837 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 6 |
Hb_000027_190 |
0.0931884885 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000044_040 |
0.0934369 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 8 |
Hb_000665_160 |
0.0949992477 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 9 |
Hb_003794_010 |
0.0955384855 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 10 |
Hb_011129_040 |
0.0962395382 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 11 |
Hb_035755_010 |
0.097730407 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_188063_020 |
0.0990611064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007645_100 |
0.0996693566 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105640858 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003939_050 |
0.100271027 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_001671_010 |
0.1008305495 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 16 |
Hb_005489_020 |
0.1009513027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005054_230 |
0.100951441 |
- |
- |
PREDICTED: wiskott-Aldrich syndrome protein family member 2 [Jatropha curcas] |
| 18 |
Hb_007441_130 |
0.1014080585 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000343_250 |
0.1026032877 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 20 |
Hb_002217_250 |
0.1029029973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |