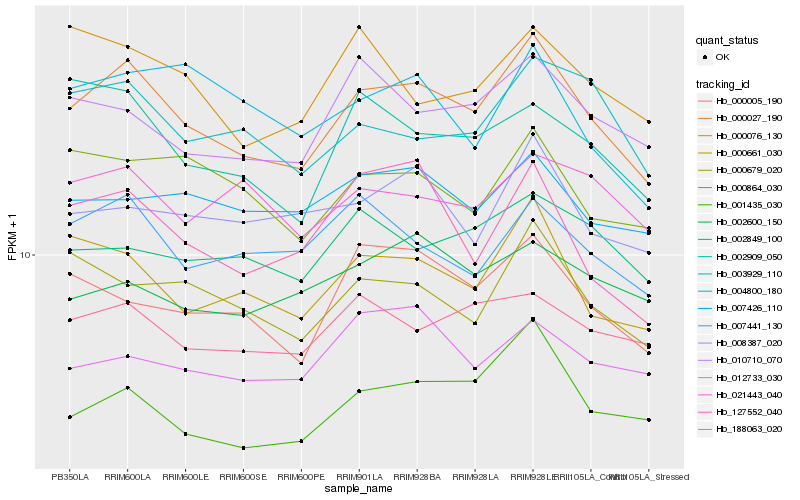
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000661_030 |
0.0787968568 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 3 |
Hb_000679_020 |
0.0870214341 |
transcription factor |
TF Family: HB |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 4 |
Hb_001435_030 |
0.0893221329 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 5 |
Hb_000005_190 |
0.0911241252 |
- |
- |
hypothetical protein JCGZ_05438 [Jatropha curcas] |
| 6 |
Hb_010710_070 |
0.0911870123 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_007441_130 |
0.0929247544 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_188063_020 |
0.0930879473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003929_110 |
0.0931884885 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000864_030 |
0.094769451 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1XR1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_008387_020 |
0.0953953797 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 12 |
Hb_007426_110 |
0.09625122 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 13 |
Hb_002849_100 |
0.0978287716 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_127552_040 |
0.0980285496 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 15 |
Hb_002909_050 |
0.0984650114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_021443_040 |
0.0992854525 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 17 |
Hb_000076_130 |
0.1042632706 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 18 |
Hb_002600_150 |
0.1045608659 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004800_180 |
0.1045960694 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 20 |
Hb_012733_030 |
0.1060803541 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |