| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
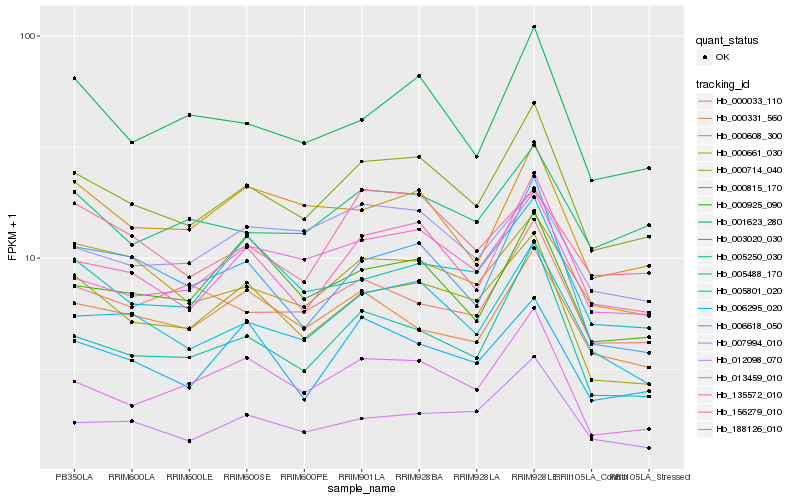
Hb_000815_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 2 |
Hb_156279_010 |
0.0526986794 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005801_020 |
0.0581060414 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 4 |
Hb_135572_010 |
0.0756102114 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 5 |
Hb_005250_030 |
0.0782743205 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 6 |
Hb_001623_280 |
0.080520604 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000714_040 |
0.0813221195 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000661_030 |
0.0841981151 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 9 |
Hb_003020_030 |
0.0848853818 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_013459_010 |
0.0875694415 |
- |
- |
PREDICTED: uncharacterized protein LOC101491556 isoform X2 [Cicer arietinum] |
| 11 |
Hb_012098_070 |
0.088462049 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 12 |
Hb_006295_020 |
0.0890336798 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 13 |
Hb_005488_170 |
0.089064668 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 14 |
Hb_006618_050 |
0.0892488149 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000608_300 |
0.0934175673 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 16 |
Hb_000925_090 |
0.0936361572 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000331_560 |
0.0936774782 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 18 |
Hb_007994_010 |
0.0937412826 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000033_110 |
0.093992816 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 20 |
Hb_188126_010 |
0.0940820483 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like isoform X3 [Jatropha curcas] |