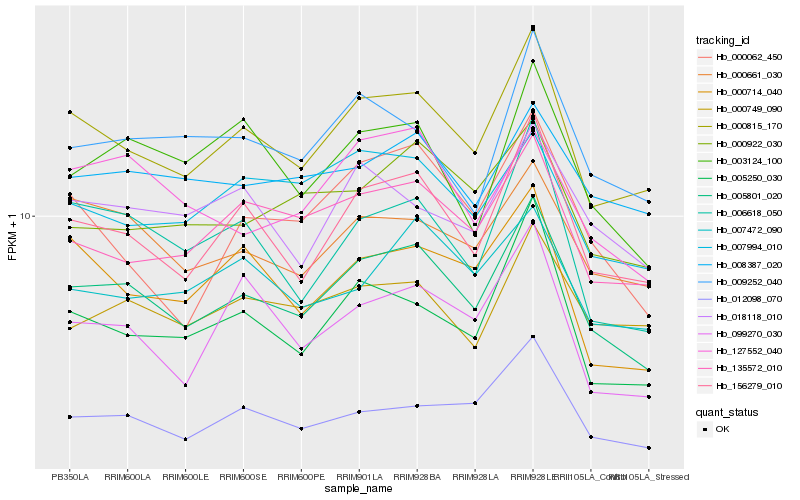
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005801_020 |
0.0 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 2 |
Hb_000815_170 |
0.0581060414 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 3 |
Hb_156279_010 |
0.0612068707 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 4 |
Hb_135572_010 |
0.0871323345 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 5 |
Hb_000661_030 |
0.089794438 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 6 |
Hb_012098_070 |
0.0903758982 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 7 |
Hb_006618_050 |
0.0903928874 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003124_100 |
0.0922738971 |
- |
- |
- |
| 9 |
Hb_005250_030 |
0.0946290924 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 10 |
Hb_000922_030 |
0.0958206508 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009252_040 |
0.0966946376 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_127552_040 |
0.0969339025 |
- |
- |
Uncharacterized protein L484_000643 [Morus notabilis] |
| 13 |
Hb_007994_010 |
0.0985456883 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008387_020 |
0.099130653 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 15 |
Hb_000749_090 |
0.0993844346 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_018118_010 |
0.1002742174 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 17 |
Hb_007472_090 |
0.1018316246 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_099270_030 |
0.1019531125 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000714_040 |
0.1024458807 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000062_450 |
0.1029459412 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |