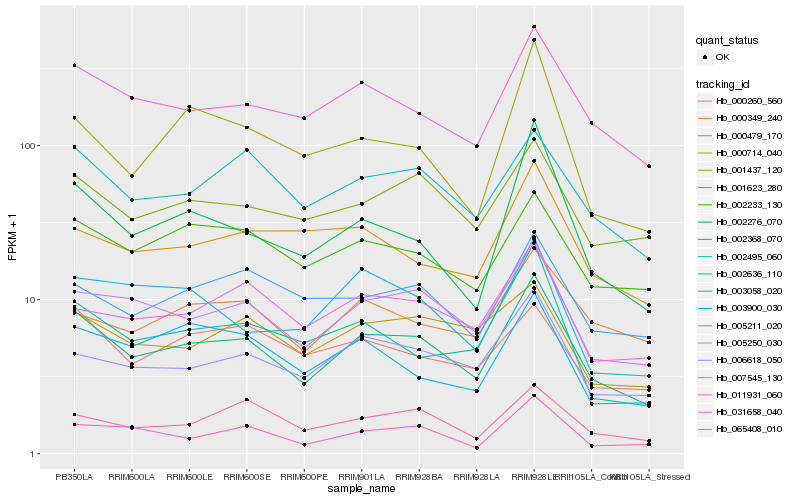
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002368_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634087 [Jatropha curcas] |
| 2 |
Hb_006618_050 |
0.0903310506 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_011931_060 |
0.0929115609 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 4 |
Hb_005250_030 |
0.1096043404 |
- |
- |
PREDICTED: uncharacterized protein LOC105640177 [Jatropha curcas] |
| 5 |
Hb_002636_110 |
0.1115911274 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 6 |
Hb_001623_280 |
0.1151940794 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_031658_040 |
0.1184540054 |
- |
- |
- |
| 8 |
Hb_002233_130 |
0.1189303467 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_007545_130 |
0.119470079 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002276_070 |
0.1200237318 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 11 |
Hb_065408_010 |
0.1228696733 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002495_060 |
0.1233068808 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_005211_020 |
0.1273608012 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000714_040 |
0.1286350883 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003900_030 |
0.1314004214 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000260_560 |
0.1331108034 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003058_020 |
0.1333140056 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000349_240 |
0.1341344105 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001437_120 |
0.1342506184 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 20 |
Hb_000479_170 |
0.1344253159 |
- |
- |
chaperone clpb, putative [Ricinus communis] |