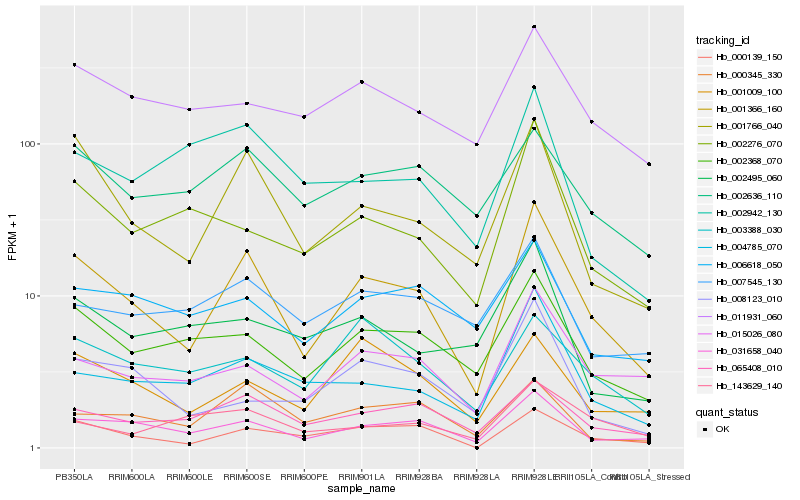
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001366_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_031658_040 |
0.1363731631 |
- |
- |
- |
| 3 |
Hb_001766_040 |
0.1394514976 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 4 |
Hb_000139_150 |
0.153981104 |
- |
- |
transposon protein, putative, Mutator sub-class [Oryza sativa Japonica Group] |
| 5 |
Hb_002368_070 |
0.1670009691 |
- |
- |
PREDICTED: uncharacterized protein LOC105634087 [Jatropha curcas] |
| 6 |
Hb_065408_010 |
0.1697126113 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 7 |
Hb_015026_080 |
0.1719193865 |
- |
- |
- |
| 8 |
Hb_004785_070 |
0.1745291232 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002276_070 |
0.1838515024 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 10 |
Hb_000345_330 |
0.1872072768 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance-like protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_001009_100 |
0.1887542671 |
- |
- |
- |
| 12 |
Hb_011931_060 |
0.1935941479 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 13 |
Hb_008123_010 |
0.193972188 |
- |
- |
- |
| 14 |
Hb_003388_030 |
0.1970012636 |
- |
- |
PREDICTED: uncharacterized protein LOC105649746 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002636_110 |
0.1978998458 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 16 |
Hb_006618_050 |
0.1999551531 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_143629_140 |
0.2048573163 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 18 |
Hb_007545_130 |
0.2052774982 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002942_130 |
0.2068464824 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002495_060 |
0.2086191364 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |