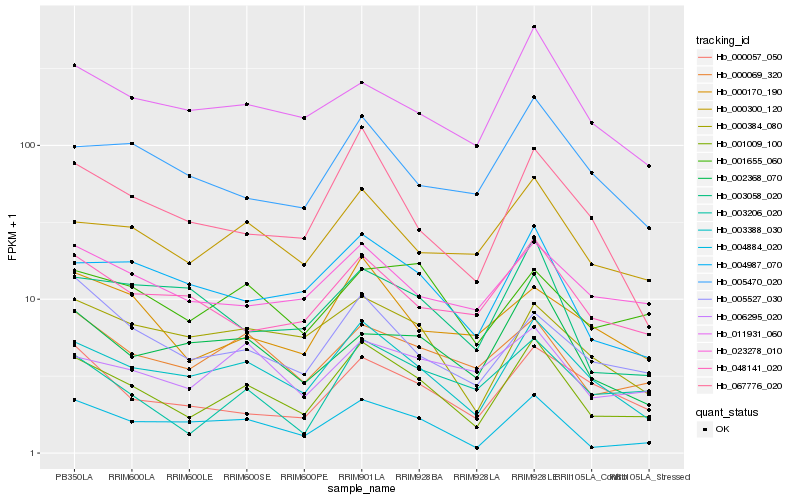
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001009_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_003388_030 |
0.0997108888 |
- |
- |
PREDICTED: uncharacterized protein LOC105649746 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004884_020 |
0.1277105494 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004987_070 |
0.1391216398 |
- |
- |
PREDICTED: heat shock 70 kDa protein 17-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000300_120 |
0.1395116803 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000069_320 |
0.1466186507 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 7 |
Hb_067776_020 |
0.1488109861 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000057_050 |
0.155262557 |
- |
- |
hypothetical protein Csa_7G132920 [Cucumis sativus] |
| 9 |
Hb_005527_030 |
0.1555330546 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g42630 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003206_020 |
0.1557039321 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 11 |
Hb_000384_080 |
0.1559931513 |
- |
- |
PREDICTED: RING-H2 finger protein ATL7-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000170_190 |
0.1587292298 |
- |
- |
PREDICTED: uncharacterized protein LOC105638988 [Jatropha curcas] |
| 13 |
Hb_011931_060 |
0.1600586132 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 14 |
Hb_048141_020 |
0.1600767092 |
- |
- |
tetratricopeptide repeat-like superfamily protein, partial [Manihot esculenta] |
| 15 |
Hb_005470_020 |
0.1622511345 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_023278_010 |
0.163146574 |
- |
- |
Chaperone protein DnaJ [Glycine soja] |
| 17 |
Hb_006295_020 |
0.1641765088 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 18 |
Hb_001655_060 |
0.1643638213 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 19 |
Hb_002368_070 |
0.1670742255 |
- |
- |
PREDICTED: uncharacterized protein LOC105634087 [Jatropha curcas] |
| 20 |
Hb_003058_020 |
0.1674962364 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |