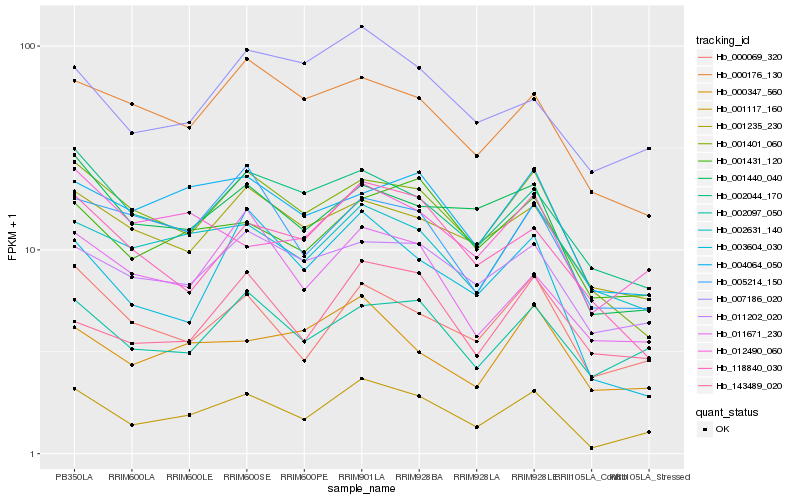
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_160 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 2 |
Hb_000069_320 |
0.1227808356 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 3 |
Hb_003604_030 |
0.1275308908 |
- |
- |
hypothetical protein MIMGU_mgv1a025764mg, partial [Erythranthe guttata] |
| 4 |
Hb_002097_050 |
0.1276818862 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 5 |
Hb_001431_120 |
0.1284227665 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 6 |
Hb_001401_060 |
0.128629176 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 7 |
Hb_001440_040 |
0.1304042193 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 8 |
Hb_011202_020 |
0.1346540118 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011671_230 |
0.1360039259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002631_140 |
0.1382593022 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002044_170 |
0.1398700107 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 12 |
Hb_004064_050 |
0.140711907 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 13 |
Hb_005214_150 |
0.1411954992 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 14 |
Hb_000347_560 |
0.142893217 |
- |
- |
- |
| 15 |
Hb_012490_060 |
0.1431837567 |
- |
- |
PREDICTED: UBP1-associated proteins 1C isoform X1 [Jatropha curcas] |
| 16 |
Hb_143489_020 |
0.1434892938 |
- |
- |
- |
| 17 |
Hb_001235_230 |
0.1441009497 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 18 |
Hb_118840_030 |
0.1442009349 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 18 homolog [Jatropha curcas] |
| 19 |
Hb_007186_020 |
0.1443265152 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 20 |
Hb_000176_130 |
0.1455593787 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |