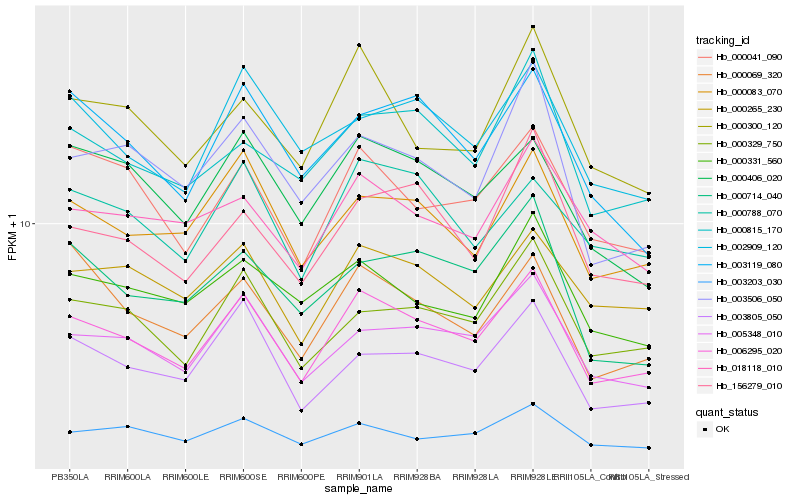
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006295_020 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 2 |
Hb_005348_010 |
0.0703171118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000083_070 |
0.0777089289 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000329_750 |
0.0780526614 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 5 |
Hb_000041_090 |
0.0802883218 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 6 |
Hb_000265_230 |
0.0802990437 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003203_030 |
0.0862943555 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 8 |
Hb_000406_020 |
0.0863403794 |
- |
- |
- |
| 9 |
Hb_003805_050 |
0.0872061086 |
- |
- |
- |
| 10 |
Hb_000788_070 |
0.0882419409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 11 |
Hb_000815_170 |
0.0890336798 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 12 |
Hb_018118_010 |
0.0908159601 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 13 |
Hb_000300_120 |
0.0908174288 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002909_120 |
0.0911250865 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 15 |
Hb_000331_560 |
0.0928735469 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 16 |
Hb_156279_010 |
0.0928793328 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003119_080 |
0.0930019373 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 18 |
Hb_003506_050 |
0.0930481859 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000069_320 |
0.0945974631 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 20 |
Hb_000714_040 |
0.094791708 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X2 [Jatropha curcas] |