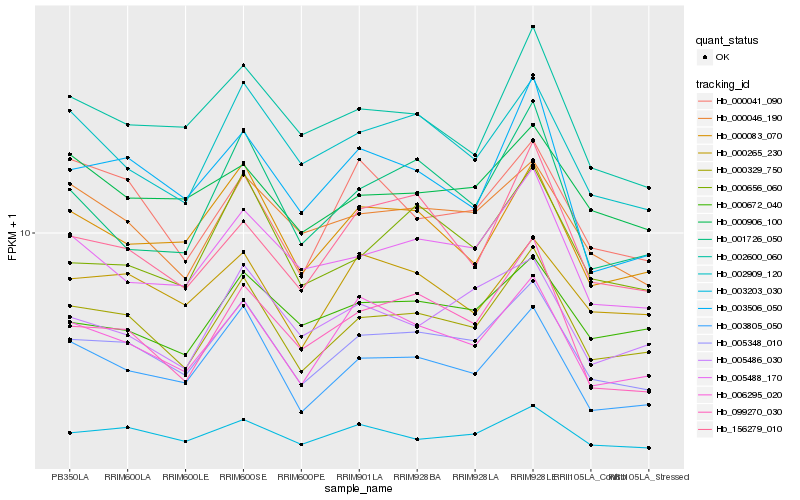
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_750 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 2 |
Hb_005348_010 |
0.0462682469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003805_050 |
0.0764786695 |
- |
- |
- |
| 4 |
Hb_006295_020 |
0.0780526614 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATR isoform X1 [Jatropha curcas] |
| 5 |
Hb_005488_170 |
0.0818335389 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 6 |
Hb_003203_030 |
0.0830356868 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 7 |
Hb_002909_120 |
0.0865282715 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 8 |
Hb_000046_190 |
0.0866569992 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 9 |
Hb_000083_070 |
0.0868447718 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002600_060 |
0.0907913068 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 11 |
Hb_000672_040 |
0.0920091665 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 12 |
Hb_000656_060 |
0.0943707706 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001726_050 |
0.0948523116 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 14 |
Hb_099270_030 |
0.0949691416 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003506_050 |
0.0956664468 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000265_230 |
0.096356754 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000041_090 |
0.0973283769 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 18 |
Hb_005486_030 |
0.0976878313 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 19 |
Hb_000906_100 |
0.0977686648 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 20 |
Hb_156279_010 |
0.0979505997 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |