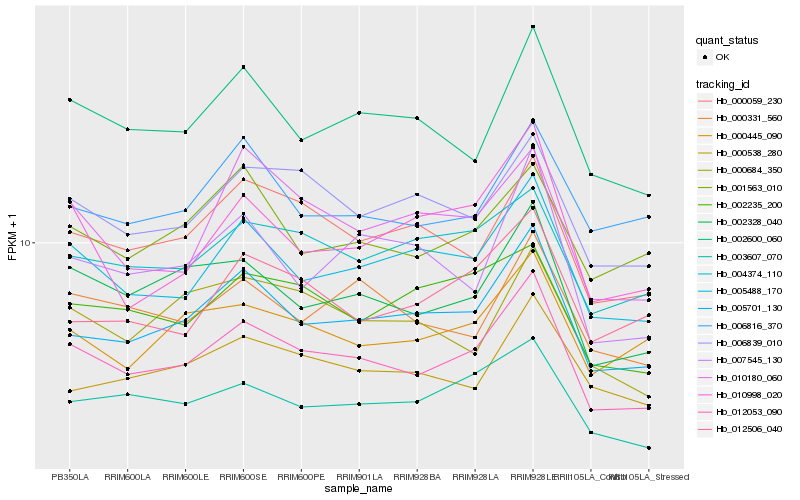
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012053_090 |
0.0 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 2 |
Hb_005701_130 |
0.0652505178 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 3 |
Hb_002328_040 |
0.0682459336 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005488_170 |
0.071177047 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 5 |
Hb_002600_060 |
0.0812707222 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 6 |
Hb_003607_070 |
0.0836447876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 7 |
Hb_000059_230 |
0.0843928778 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 8 |
Hb_006839_010 |
0.0869208441 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 9 |
Hb_000331_560 |
0.0910432471 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 10 |
Hb_000445_090 |
0.0924876839 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 11 |
Hb_004374_110 |
0.0933161088 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 12 |
Hb_010998_020 |
0.0977005129 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 13 |
Hb_007545_130 |
0.0977396406 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000538_280 |
0.0977735266 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 15 |
Hb_010180_060 |
0.0983419999 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_012506_040 |
0.0991951714 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 17 |
Hb_002235_200 |
0.0992784353 |
- |
- |
pattern formation protein, putative [Ricinus communis] |
| 18 |
Hb_000684_350 |
0.0997321465 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001563_010 |
0.1012107123 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 20 |
Hb_006816_370 |
0.1017213251 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |