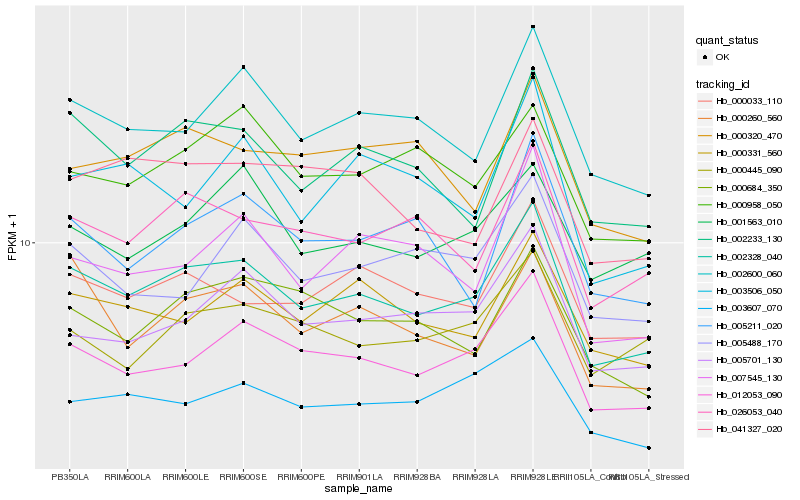
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002328_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 2 |
Hb_012053_090 |
0.0682459336 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 3 |
Hb_002233_130 |
0.081917604 |
- |
- |
transporter, putative [Ricinus communis] |
| 4 |
Hb_002600_060 |
0.0820581084 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 5 |
Hb_026053_040 |
0.0830146673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000033_110 |
0.087302981 |
- |
- |
hypothetical protein JCGZ_19253 [Jatropha curcas] |
| 7 |
Hb_000331_560 |
0.0893360866 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 8 |
Hb_005701_130 |
0.0938798082 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 9 |
Hb_003607_070 |
0.09665521 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910 [Jatropha curcas] |
| 10 |
Hb_000260_560 |
0.0976024161 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000684_350 |
0.0980972397 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007545_130 |
0.1001144543 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000445_090 |
0.1005865849 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 14 |
Hb_000320_470 |
0.1018888115 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 15 |
Hb_001563_010 |
0.1021167979 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 16 |
Hb_000958_050 |
0.1026370354 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 17 |
Hb_005488_170 |
0.1032082771 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 18 |
Hb_003506_050 |
0.1035631413 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_005211_020 |
0.1038643452 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_041327_020 |
0.1048212356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |