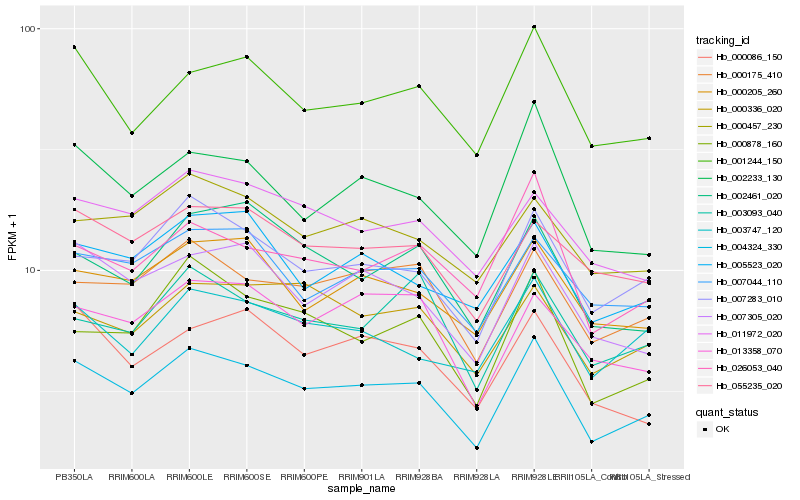
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_330 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 2 |
Hb_007283_010 |
0.0695053365 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 3 |
Hb_000175_410 |
0.0740394143 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002233_130 |
0.0743045751 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_007305_020 |
0.0771104028 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 6 |
Hb_001244_150 |
0.0771134507 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 7 |
Hb_000205_260 |
0.0827036547 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 8 |
Hb_000878_160 |
0.0829178909 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 9 |
Hb_026053_040 |
0.0829326183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007044_110 |
0.0862721798 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_055235_020 |
0.0867846332 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003747_120 |
0.087474407 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 13 |
Hb_011972_020 |
0.087628364 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_005523_020 |
0.0877270664 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 15 |
Hb_000086_150 |
0.0880432303 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002461_020 |
0.088503668 |
- |
- |
PREDICTED: uncharacterized protein LOC105642649 isoform X2 [Jatropha curcas] |
| 17 |
Hb_013358_070 |
0.0888659741 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000457_230 |
0.0891098039 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000336_020 |
0.0894041803 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 20 |
Hb_003093_040 |
0.090224227 |
- |
- |
PREDICTED: nucleolar complex protein 4 homolog isoform X1 [Jatropha curcas] |