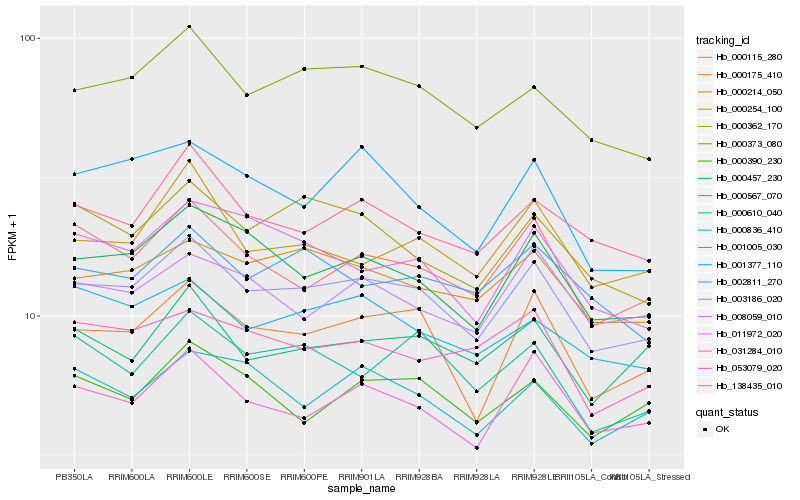
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003186_020 |
0.0 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000373_080 |
0.0491074424 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 3 |
Hb_000457_230 |
0.0517199038 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000175_410 |
0.053599513 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_031284_010 |
0.0548816107 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 6 |
Hb_000214_050 |
0.0558117822 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000115_280 |
0.0562598408 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 8 |
Hb_138435_010 |
0.0588475396 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 9 |
Hb_000254_100 |
0.0632690908 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 10 |
Hb_001005_030 |
0.0637412685 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000362_170 |
0.0642831772 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 12 |
Hb_008059_010 |
0.0662821766 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000836_410 |
0.0664702447 |
- |
- |
sec10, putative [Ricinus communis] |
| 14 |
Hb_002811_270 |
0.0665355722 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 15 |
Hb_011972_020 |
0.0667193641 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_000567_070 |
0.0671265458 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 17 |
Hb_001377_110 |
0.0677887159 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 18 |
Hb_000610_040 |
0.0680539543 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000390_230 |
0.0684881522 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 20 |
Hb_053079_020 |
0.0690099346 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |