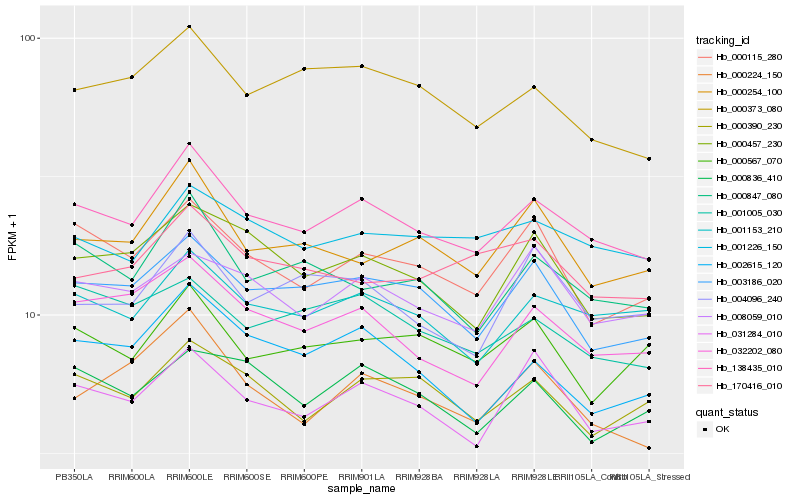
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138435_010 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 2 |
Hb_001153_210 |
0.0529780313 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003186_020 |
0.0588475396 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031284_010 |
0.0620564167 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 5 |
Hb_000847_080 |
0.0623743648 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_032202_080 |
0.062731554 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001226_150 |
0.0642198898 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 8 |
Hb_000115_280 |
0.0662545327 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_000373_080 |
0.0664274549 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 10 |
Hb_000390_230 |
0.0667705446 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 11 |
Hb_000457_230 |
0.067517258 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008059_010 |
0.0677718509 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000254_100 |
0.0680469416 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 14 |
Hb_001005_030 |
0.0701540925 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_002615_120 |
0.0727662187 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000224_150 |
0.0736280126 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000836_410 |
0.0739245403 |
- |
- |
sec10, putative [Ricinus communis] |
| 18 |
Hb_000567_070 |
0.0752286778 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 19 |
Hb_170416_010 |
0.0753746279 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 20 |
Hb_004096_240 |
0.0754482819 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |