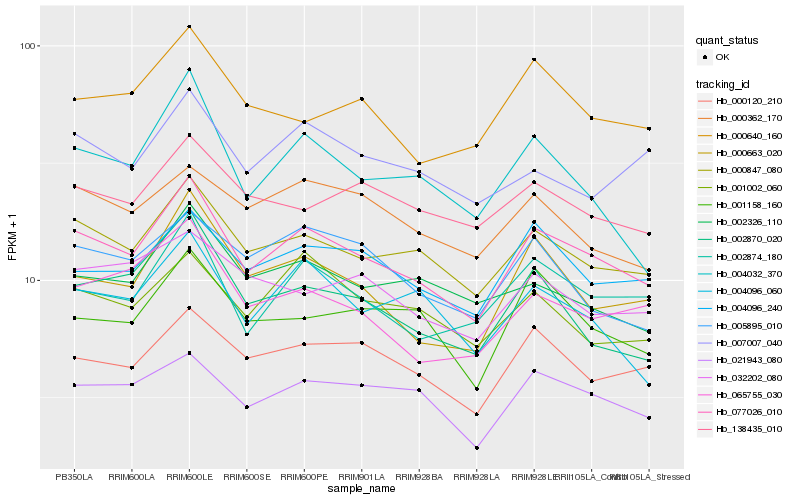
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077026_010 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 2 |
Hb_000847_080 |
0.0566274939 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 3 |
Hb_002874_180 |
0.0722656609 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_021943_080 |
0.0730277766 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000120_210 |
0.0779159413 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 6 |
Hb_004096_240 |
0.0780336638 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 7 |
Hb_000663_020 |
0.0782206559 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_065755_030 |
0.0869817059 |
- |
- |
- |
| 9 |
Hb_001158_160 |
0.0871288187 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002870_020 |
0.0880217283 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000640_160 |
0.088642644 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 12 |
Hb_001002_060 |
0.0896210314 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 13 |
Hb_138435_010 |
0.089710105 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 14 |
Hb_000362_170 |
0.0901116571 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_004032_370 |
0.0904037646 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 16 |
Hb_004096_060 |
0.0904097152 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 17 |
Hb_002326_110 |
0.0919069878 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 18 |
Hb_032202_080 |
0.0924835393 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005895_010 |
0.0936296459 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007007_040 |
0.0937610648 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |