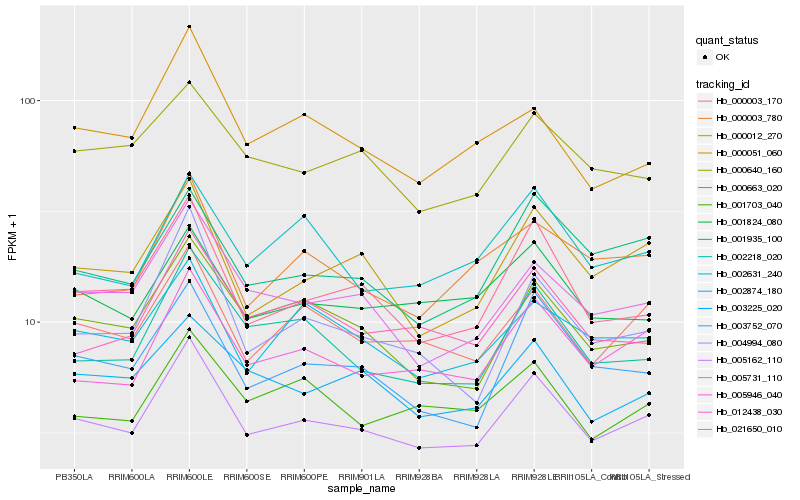
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005162_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 2 |
Hb_005946_040 |
0.0654645205 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 3 |
Hb_021650_010 |
0.0663594269 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 4 |
Hb_000003_170 |
0.0688477827 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 5 |
Hb_005731_110 |
0.0741805389 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 6 |
Hb_000051_060 |
0.0742046269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003752_070 |
0.0759709477 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002218_020 |
0.0782853975 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 9 |
Hb_000663_020 |
0.0794655909 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_004994_080 |
0.079519902 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001935_100 |
0.0823410834 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 12 |
Hb_012438_030 |
0.0838713456 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 13 |
Hb_002874_180 |
0.0853908077 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001824_080 |
0.0870939814 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase isoform X1 [Jatropha curcas] |
| 15 |
Hb_001703_040 |
0.0881149417 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000012_270 |
0.0885717093 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 17 |
Hb_000003_780 |
0.0887818268 |
- |
- |
hexokinase [Manihot esculenta] |
| 18 |
Hb_000640_160 |
0.0897144679 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 19 |
Hb_002631_240 |
0.0901647724 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 20 |
Hb_003225_020 |
0.0902073727 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |