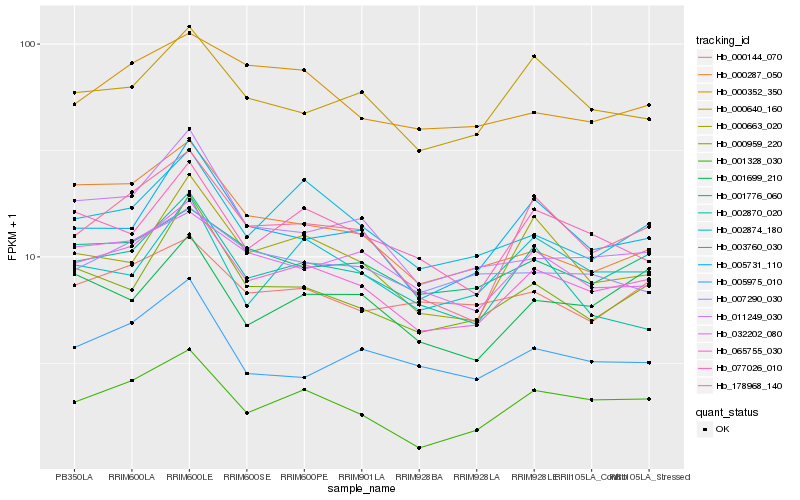
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065755_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_178968_140 |
0.0707997624 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_001328_030 |
0.0723963137 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 4 |
Hb_003760_030 |
0.0775451844 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 5 |
Hb_005731_110 |
0.0792856499 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 6 |
Hb_000959_220 |
0.0811543217 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_001776_060 |
0.0812236309 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 8 |
Hb_000287_050 |
0.0866539842 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 9 |
Hb_032202_080 |
0.0867225755 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_077026_010 |
0.0869817059 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 11 |
Hb_000640_160 |
0.0877095851 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 12 |
Hb_011249_030 |
0.088610613 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 13 |
Hb_000663_020 |
0.0887863989 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002870_020 |
0.0932414347 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000352_350 |
0.0942809335 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005975_010 |
0.0943287505 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000144_070 |
0.0947134189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001699_210 |
0.0952834444 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007290_030 |
0.0978927721 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 20 |
Hb_002874_180 |
0.0984879111 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |