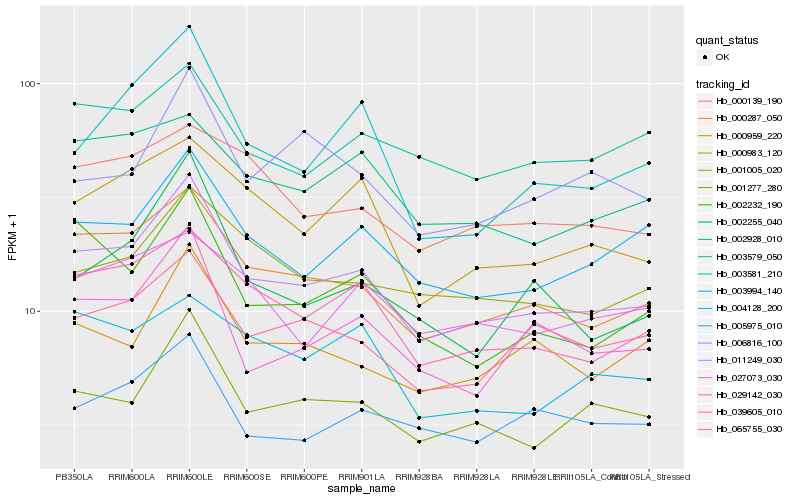
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011249_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 2 |
Hb_000287_050 |
0.0564565556 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 3 |
Hb_039605_010 |
0.074592849 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001277_280 |
0.078563539 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 5 |
Hb_004128_200 |
0.0832299917 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 6 |
Hb_029142_030 |
0.0873611279 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 7 |
Hb_065755_030 |
0.088610613 |
- |
- |
- |
| 8 |
Hb_000983_120 |
0.0921888698 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000959_220 |
0.0933220632 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_002232_190 |
0.0936724816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001005_020 |
0.0964443335 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 12 |
Hb_002928_010 |
0.0969858568 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 13 |
Hb_003581_210 |
0.0971383474 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002255_040 |
0.0985064807 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 15 |
Hb_005975_010 |
0.0995998152 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000139_190 |
0.1012906038 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003579_050 |
0.10357436 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 18 |
Hb_027073_030 |
0.1071031621 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 19 |
Hb_006816_100 |
0.1075921185 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 20 |
Hb_003994_140 |
0.1079045778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |