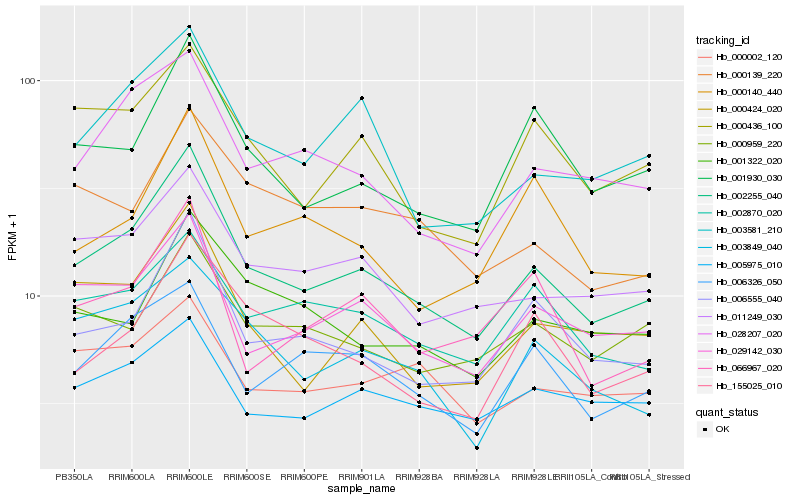
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002255_040 |
0.0 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 2 |
Hb_006555_040 |
0.0914931129 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 3 |
Hb_029142_030 |
0.0977666544 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 4 |
Hb_011249_030 |
0.0985064807 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 5 |
Hb_028207_020 |
0.1040718891 |
- |
- |
PREDICTED: uncharacterized protein LOC105650315 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003581_210 |
0.1043153579 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000424_020 |
0.1048879352 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 8 |
Hb_001930_030 |
0.1072393175 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000436_100 |
0.1089997121 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 10 |
Hb_006326_050 |
0.1128721889 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_005975_010 |
0.1147680758 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003849_040 |
0.1156564715 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 13 |
Hb_155025_010 |
0.1179908504 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 14 |
Hb_002870_020 |
0.1180692727 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000959_220 |
0.1194975057 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000139_220 |
0.1212316705 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_001322_020 |
0.1220228599 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000002_120 |
0.122598855 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 19 |
Hb_000140_440 |
0.1227705412 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 20 |
Hb_066967_020 |
0.1234371576 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |