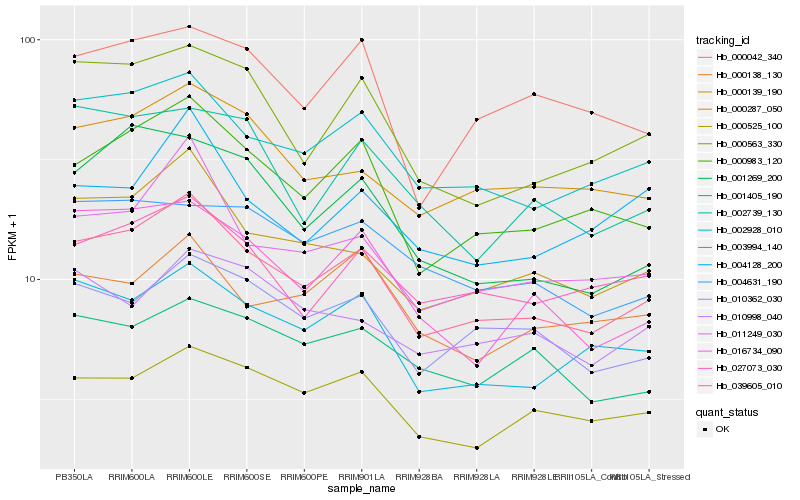
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039605_010 |
0.0 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002928_010 |
0.0546425686 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 3 |
Hb_000983_120 |
0.0581990807 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000287_050 |
0.0646479734 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 5 |
Hb_003994_140 |
0.0722944188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011249_030 |
0.074592849 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 7 |
Hb_000563_330 |
0.0772948648 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000525_100 |
0.077380853 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 9 |
Hb_000139_190 |
0.0779254529 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_027073_030 |
0.0797083573 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 11 |
Hb_010362_030 |
0.0834653099 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_016734_090 |
0.0884952058 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 13 |
Hb_001269_200 |
0.0924725367 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 14 |
Hb_000042_340 |
0.0938150844 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 15 |
Hb_001405_190 |
0.0973190152 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_004631_190 |
0.0974967265 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 17 |
Hb_004128_200 |
0.0980875369 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 18 |
Hb_010998_040 |
0.0983222469 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 19 |
Hb_002739_130 |
0.0987780846 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 20 |
Hb_000138_130 |
0.0991872597 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |