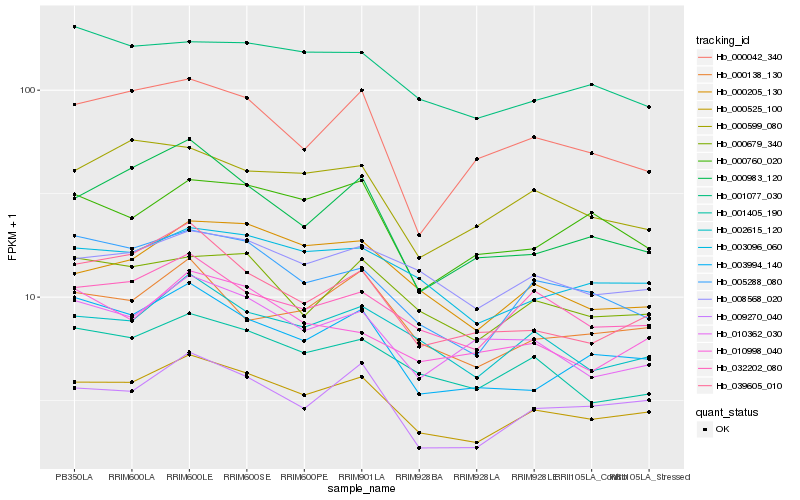
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000525_100 |
0.0 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 2 |
Hb_009270_040 |
0.0653634302 |
- |
- |
- |
| 3 |
Hb_005288_080 |
0.0681784469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002615_120 |
0.0693586386 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003096_060 |
0.0697300292 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 6 |
Hb_032202_080 |
0.073298201 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000679_340 |
0.0750739895 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001405_190 |
0.0755779308 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000205_130 |
0.075873568 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_003994_140 |
0.0764298281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_039605_010 |
0.077380853 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000138_130 |
0.0783949687 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 13 |
Hb_008568_020 |
0.0805175052 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 14 |
Hb_000599_080 |
0.0820294698 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 15 |
Hb_000983_120 |
0.0832983333 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_010362_030 |
0.0837690993 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_001077_030 |
0.0859785825 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000042_340 |
0.0865614031 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 19 |
Hb_000760_020 |
0.086924603 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_010998_040 |
0.0879911874 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |