| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
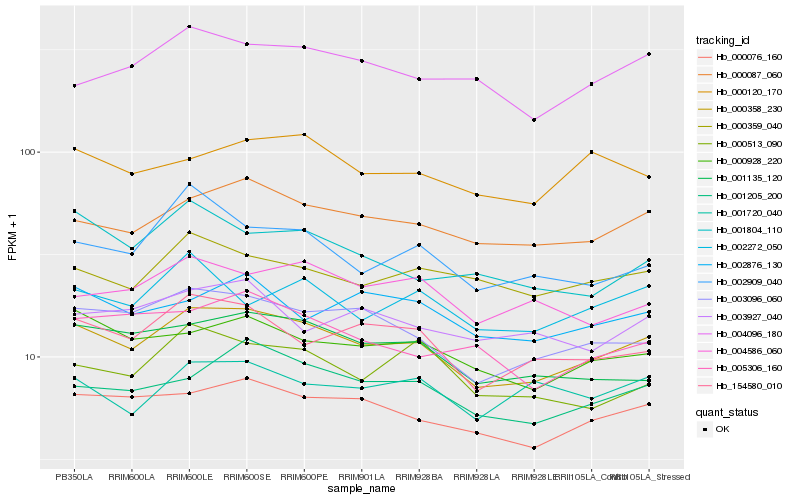
Hb_000358_230 |
0.0 |
- |
- |
protein phosphatases pp1 regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_000087_060 |
0.0577885045 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 3 |
Hb_000076_160 |
0.0595278364 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 4 |
Hb_003096_060 |
0.0627926885 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 5 |
Hb_001205_200 |
0.0670575165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_154580_010 |
0.0680318036 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 7 |
Hb_000928_220 |
0.0694561968 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 8 |
Hb_001135_120 |
0.0759904916 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000359_040 |
0.0775869341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004096_180 |
0.0797599281 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 11 |
Hb_002876_130 |
0.0801691171 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 12 |
Hb_000120_170 |
0.080428184 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 13 |
Hb_005306_160 |
0.0811916595 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_004586_060 |
0.0814706205 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_002909_040 |
0.081806833 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 16 |
Hb_002272_050 |
0.0821376545 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 17 |
Hb_000513_090 |
0.0822790049 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 18 |
Hb_001720_040 |
0.0838661685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001804_110 |
0.0842997555 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 20 |
Hb_003927_040 |
0.084723071 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |