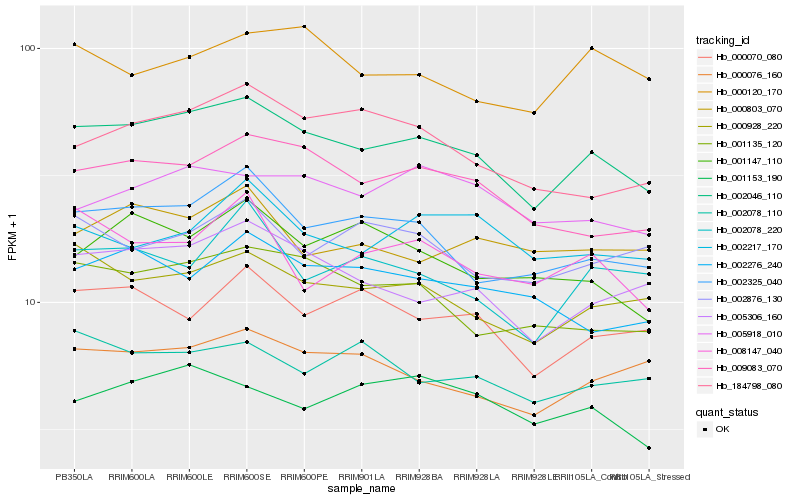
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002046_110 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |
| 2 |
Hb_009083_070 |
0.0659400237 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 3 |
Hb_002325_040 |
0.0665278339 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005306_160 |
0.0668702375 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_000928_220 |
0.0671846272 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 6 |
Hb_001135_120 |
0.0690483227 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000076_160 |
0.0760201609 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 8 |
Hb_000070_080 |
0.0782271007 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 9 |
Hb_184798_080 |
0.0787764839 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 10 |
Hb_005918_010 |
0.079774769 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3-like [Jatropha curcas] |
| 11 |
Hb_001153_190 |
0.0816145732 |
- |
- |
PREDICTED: DNA-directed RNA polymerase IV subunit 1 [Jatropha curcas] |
| 12 |
Hb_001147_110 |
0.0838629869 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 13 |
Hb_002276_240 |
0.0838893721 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 14 |
Hb_002078_220 |
0.0843924805 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002078_110 |
0.0849449365 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 16 |
Hb_000120_170 |
0.0849540305 |
- |
- |
selenoprotein [Populus trichocarpa] |
| 17 |
Hb_002217_170 |
0.0861645617 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL6-like isoform X1 [Gossypium raimondii] |
| 18 |
Hb_000803_070 |
0.0866850703 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 19 |
Hb_002876_130 |
0.0868560178 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 20 |
Hb_008147_040 |
0.0869808475 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |