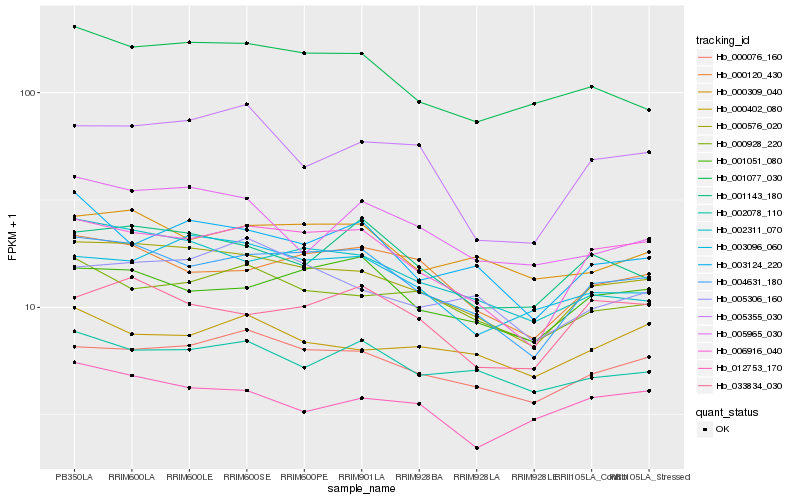
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000576_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 2 |
Hb_004631_180 |
0.0517988293 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 3 |
Hb_006916_040 |
0.0591670372 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 4 |
Hb_000076_160 |
0.061643544 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 5 |
Hb_002311_070 |
0.0652146305 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 6 |
Hb_000928_220 |
0.0683204325 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 7 |
Hb_001077_030 |
0.0734758996 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003096_060 |
0.0741253444 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 9 |
Hb_005306_160 |
0.0757278046 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_000120_430 |
0.076428373 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 11 |
Hb_001143_180 |
0.0766303564 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 12 |
Hb_000309_040 |
0.0786978013 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 13 |
Hb_005355_030 |
0.0794950933 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003124_220 |
0.0795461165 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 15 |
Hb_005965_030 |
0.0795822652 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 16 |
Hb_002078_110 |
0.0802183707 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 17 |
Hb_033834_030 |
0.0809709435 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_012753_170 |
0.0821191347 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001051_080 |
0.0826628228 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 20 |
Hb_000402_080 |
0.0848585065 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |