| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
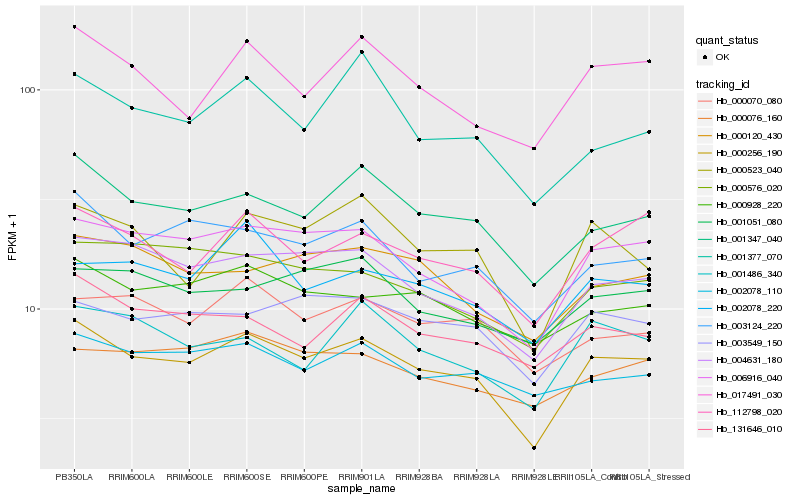
Hb_000256_190 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 2 |
Hb_006916_040 |
0.0619038903 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 3 |
Hb_001347_040 |
0.0670116677 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 4 |
Hb_004631_180 |
0.0688032917 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 5 |
Hb_112798_020 |
0.0740368063 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 6 |
Hb_003124_220 |
0.0761700653 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 7 |
Hb_003549_150 |
0.0808780863 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 8 |
Hb_017491_030 |
0.0809990334 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 9 |
Hb_000928_220 |
0.0829481483 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 10 |
Hb_000523_040 |
0.0839101114 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000120_430 |
0.0863513956 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 12 |
Hb_131646_010 |
0.0874173768 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000070_080 |
0.0879366895 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001486_340 |
0.0887360391 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_002078_220 |
0.0888607693 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000576_020 |
0.0888790082 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 17 |
Hb_000076_160 |
0.0894596694 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 18 |
Hb_002078_110 |
0.0897856881 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 19 |
Hb_001051_080 |
0.0903761807 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 20 |
Hb_001377_070 |
0.0921729943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |