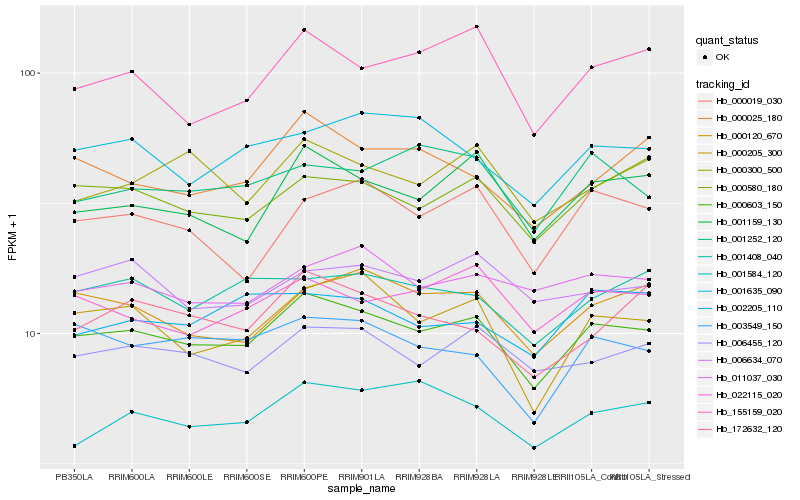
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000603_150 |
0.0 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 2 |
Hb_006455_120 |
0.058516659 |
- |
- |
n6-DNA-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001159_130 |
0.0603912825 |
- |
- |
PREDICTED: protein cornichon homolog 4 [Jatropha curcas] |
| 4 |
Hb_000205_300 |
0.0615924966 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 5 |
Hb_000120_670 |
0.0626868379 |
- |
- |
PREDICTED: SRSF protein kinase 2-like [Jatropha curcas] |
| 6 |
Hb_000580_180 |
0.0630336512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003549_150 |
0.0637241588 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 8 |
Hb_022115_020 |
0.0657129146 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001408_040 |
0.0659023183 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001584_120 |
0.0666706895 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase 3 [Jatropha curcas] |
| 11 |
Hb_011037_030 |
0.068588142 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 12 |
Hb_000025_180 |
0.0686991327 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 13 |
Hb_000300_500 |
0.0687345101 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 14 |
Hb_000019_030 |
0.0692314109 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 15 |
Hb_155159_020 |
0.0697135334 |
- |
- |
PREDICTED: clathrin light chain 1-like [Jatropha curcas] |
| 16 |
Hb_001635_090 |
0.0700696677 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 17 |
Hb_172632_120 |
0.0708838164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001252_120 |
0.0713795973 |
- |
- |
PREDICTED: delta-1-pyrroline-5-carboxylate synthase-like [Gossypium raimondii] |
| 19 |
Hb_006634_070 |
0.071958031 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 20 |
Hb_002205_110 |
0.0730964838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |