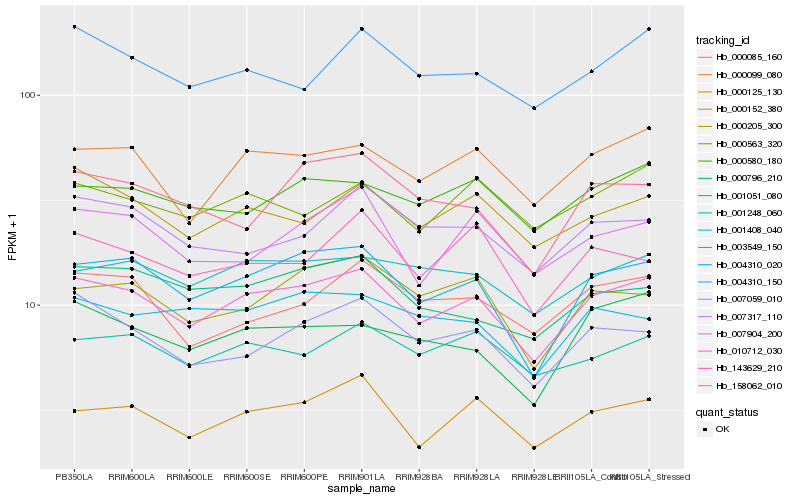
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010712_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004310_020 |
0.0490264214 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000099_080 |
0.0614094818 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 4 |
Hb_000125_130 |
0.0645098675 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC104879785 [Vitis vinifera] |
| 5 |
Hb_000152_380 |
0.0698083682 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 6 |
Hb_001408_040 |
0.0707013071 |
- |
- |
PREDICTED: MACPF domain-containing protein At4g24290 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007059_010 |
0.0709220776 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |
| 8 |
Hb_001248_060 |
0.071878578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000205_300 |
0.071960034 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 10 |
Hb_000580_180 |
0.0737482223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001051_080 |
0.0763700267 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 12 |
Hb_004310_150 |
0.0772027122 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 13 |
Hb_000563_320 |
0.0785175509 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 14 |
Hb_007904_200 |
0.0787691742 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003549_150 |
0.0796005709 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 16 |
Hb_000796_210 |
0.0797528971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000085_160 |
0.0798449774 |
- |
- |
hypothetical protein CICLE_v10009012mg [Citrus clementina] |
| 18 |
Hb_158062_010 |
0.0800618124 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 19 |
Hb_007317_110 |
0.0803180458 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C660.15-like [Jatropha curcas] |
| 20 |
Hb_143629_210 |
0.0803199799 |
- |
- |
PREDICTED: transcription factor PIF3 isoform X1 [Jatropha curcas] |