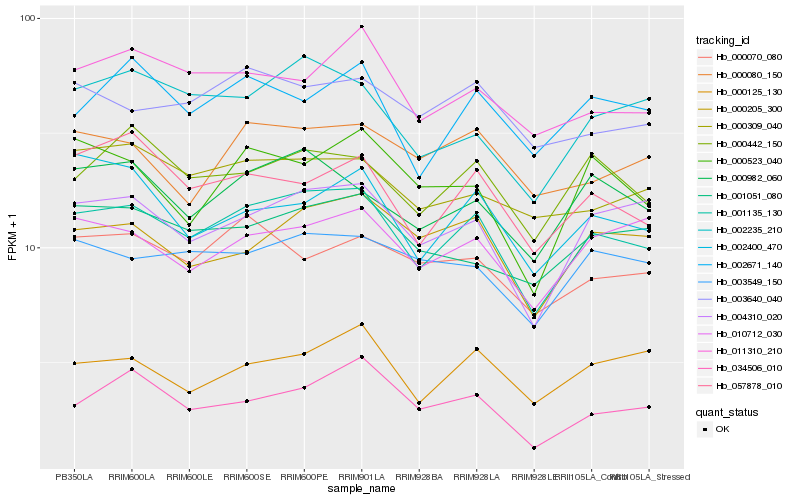
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001135_130 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 2 |
Hb_004310_020 |
0.0737253351 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000982_060 |
0.0811837509 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 4 |
Hb_000442_150 |
0.0834572522 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 5 |
Hb_000205_300 |
0.0853793885 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 6 |
Hb_002235_210 |
0.0854907449 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 7 |
Hb_057878_010 |
0.0860977695 |
- |
- |
hypothetical protein VITISV_036304 [Vitis vinifera] |
| 8 |
Hb_011310_210 |
0.0881713884 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 9 |
Hb_010712_030 |
0.0882257255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003549_150 |
0.0888357956 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 11 |
Hb_002671_140 |
0.0902928512 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 12 |
Hb_001051_080 |
0.0912367778 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 13 |
Hb_003640_040 |
0.0924016137 |
- |
- |
PREDICTED: uncharacterized protein LOC105647348 [Jatropha curcas] |
| 14 |
Hb_000309_040 |
0.0926700669 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 15 |
Hb_000125_130 |
0.0934852304 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC104879785 [Vitis vinifera] |
| 16 |
Hb_034506_010 |
0.0936969523 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 17 |
Hb_000070_080 |
0.0951614401 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000080_150 |
0.0961086407 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000523_040 |
0.0962652511 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002400_470 |
0.0968613978 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial isoform X1 [Jatropha curcas] |