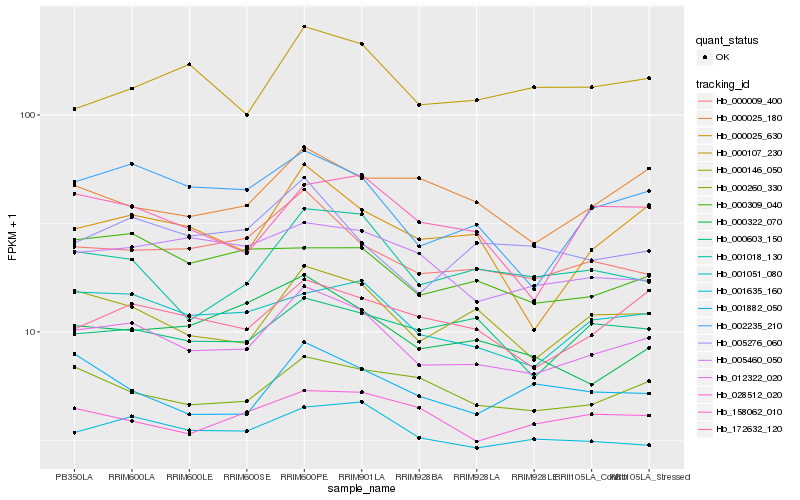
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012322_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000146_050 |
0.0644198327 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 3 |
Hb_000009_400 |
0.0721484229 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 4 |
Hb_001051_080 |
0.0722580145 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 5 |
Hb_001635_160 |
0.073336992 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 6 |
Hb_172632_120 |
0.0782064338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002235_210 |
0.0789087093 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 8 |
Hb_000260_330 |
0.0801578837 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |
| 9 |
Hb_001018_130 |
0.0837653769 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 10 |
Hb_000025_630 |
0.0869795787 |
- |
- |
PREDICTED: uncharacterized protein LOC105635952 [Jatropha curcas] |
| 11 |
Hb_005460_050 |
0.0897045704 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 12 |
Hb_005276_060 |
0.0897841475 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000107_230 |
0.0898529601 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_028512_020 |
0.0907536322 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 15 |
Hb_158062_010 |
0.0912381456 |
- |
- |
PREDICTED: uncharacterized protein LOC105634413 [Jatropha curcas] |
| 16 |
Hb_000025_180 |
0.0914791728 |
- |
- |
hypothetical protein CISIN_1g024241mg [Citrus sinensis] |
| 17 |
Hb_001882_050 |
0.0918963173 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000603_150 |
0.0920826641 |
- |
- |
PREDICTED: guanine nucleotide-binding protein alpha-1 subunit [Jatropha curcas] |
| 19 |
Hb_000309_040 |
0.0920964107 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 20 |
Hb_000322_070 |
0.092914321 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |