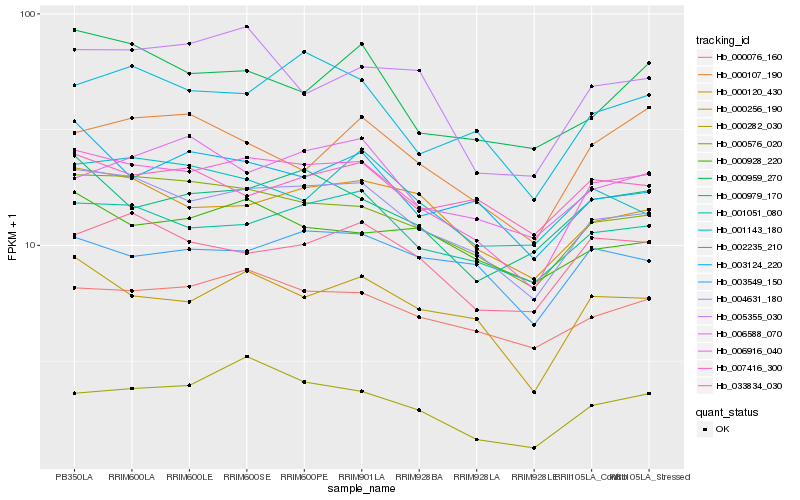
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_040 |
0.0 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 2 |
Hb_004631_180 |
0.0388647305 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 3 |
Hb_000576_020 |
0.0591670372 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 4 |
Hb_000256_190 |
0.0619038903 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001051_080 |
0.0691616236 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 6 |
Hb_000076_160 |
0.0697364028 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 7 |
Hb_033834_030 |
0.0760002681 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_430 |
0.076159027 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 9 |
Hb_002235_210 |
0.0794634914 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 10 |
Hb_000979_170 |
0.079992284 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 11 |
Hb_000282_030 |
0.0845281788 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 12 |
Hb_003549_150 |
0.0846461191 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 13 |
Hb_003124_220 |
0.0860535502 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 14 |
Hb_000928_220 |
0.0887227992 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 15 |
Hb_005355_030 |
0.0893789722 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001143_180 |
0.0905256374 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 17 |
Hb_000107_190 |
0.0912367946 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 18 |
Hb_007416_300 |
0.0915758051 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 19 |
Hb_000959_270 |
0.0916063075 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 20 |
Hb_006588_070 |
0.0917678192 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |