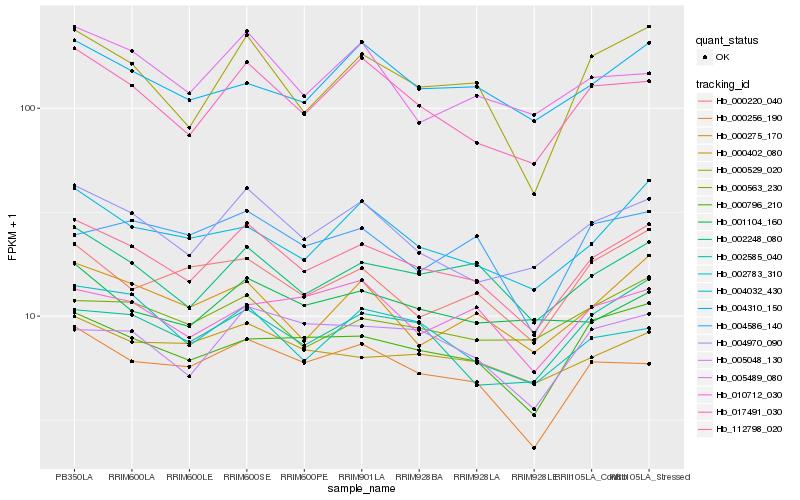
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112798_020 |
0.0 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 2 |
Hb_002248_080 |
0.0603352096 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004970_090 |
0.0620817198 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 4 |
Hb_017491_030 |
0.0638633579 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 5 |
Hb_000529_020 |
0.0676225274 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 6 |
Hb_000796_210 |
0.0733368032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000256_190 |
0.0740368063 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000402_080 |
0.0768967659 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 9 |
Hb_004032_430 |
0.0769839761 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 10 |
Hb_005048_130 |
0.0771049219 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 11 |
Hb_000275_170 |
0.0783554228 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 12 |
Hb_002585_040 |
0.0795517196 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 13 |
Hb_000563_230 |
0.0841583882 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 14 |
Hb_004310_150 |
0.0848960324 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 15 |
Hb_002783_310 |
0.0850117189 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 16 |
Hb_000220_040 |
0.0859789033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010712_030 |
0.0883381904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001104_160 |
0.0885387581 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 19 |
Hb_005489_080 |
0.0890574873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004586_140 |
0.0902591792 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |