| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
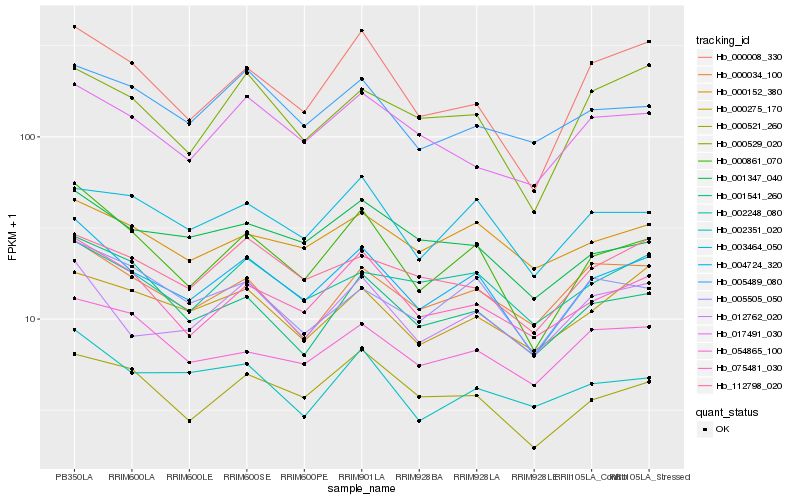
Hb_004724_320 |
0.0 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 2 |
Hb_000861_070 |
0.0500093152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_075481_030 |
0.0724629927 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 4 |
Hb_012762_020 |
0.0763099872 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 5 |
Hb_000008_330 |
0.0822544714 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 6 |
Hb_000521_260 |
0.0849129318 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_005505_050 |
0.0864014326 |
- |
- |
PREDICTED: uncharacterized protein LOC105639309 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002248_080 |
0.0865778711 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000034_100 |
0.0868975672 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 10 |
Hb_003464_050 |
0.089802439 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 11 |
Hb_112798_020 |
0.0904534168 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 12 |
Hb_000529_020 |
0.0913607549 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 13 |
Hb_000275_170 |
0.0916799144 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 14 |
Hb_000152_380 |
0.091940158 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 15 |
Hb_054865_100 |
0.0920803946 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001347_040 |
0.0924319017 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 17 |
Hb_001541_260 |
0.0943488477 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 18 |
Hb_017491_030 |
0.0945213876 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 19 |
Hb_002351_020 |
0.0947297479 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 20 |
Hb_005489_080 |
0.09536345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |