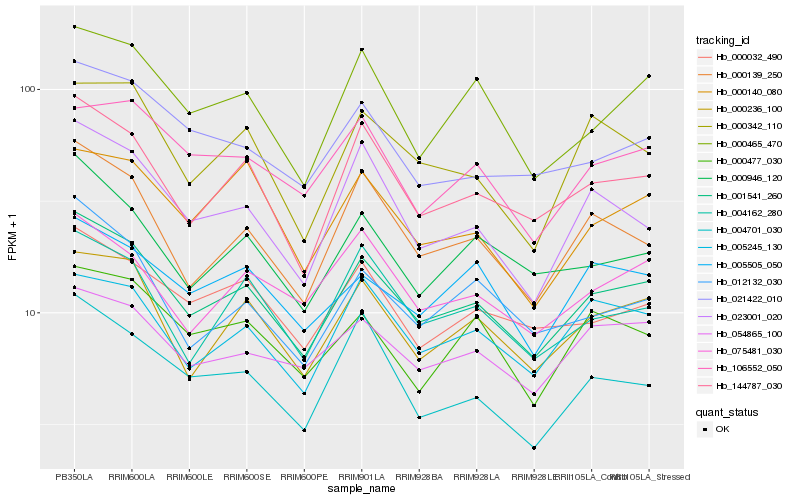
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_260 |
0.0 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 2 |
Hb_144787_030 |
0.05881462 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000236_100 |
0.0645865562 |
- |
- |
Dynein alpha chain, flagellar outer arm [Gossypium arboreum] |
| 4 |
Hb_000139_250 |
0.0686460042 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 5 |
Hb_000465_470 |
0.0721124341 |
- |
- |
unknown [Picea sitchensis] |
| 6 |
Hb_000032_490 |
0.0730335244 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 7 |
Hb_000946_120 |
0.0743757818 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_023001_020 |
0.0753213335 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 9 |
Hb_004162_280 |
0.0759208573 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 10 |
Hb_054865_100 |
0.0767122426 |
- |
- |
PREDICTED: probable protein S-acyltransferase 12 isoform X1 [Jatropha curcas] |
| 11 |
Hb_021422_010 |
0.0785461856 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_005245_130 |
0.0801379905 |
- |
- |
PREDICTED: uncharacterized protein LOC105643730 [Jatropha curcas] |
| 13 |
Hb_012132_030 |
0.0803905656 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 14 |
Hb_075481_030 |
0.0825902611 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000342_110 |
0.082962786 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 16 |
Hb_000477_030 |
0.0833087867 |
- |
- |
PREDICTED: boron transporter 4-like [Jatropha curcas] |
| 17 |
Hb_004701_030 |
0.0838190743 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 18 |
Hb_000140_080 |
0.0838260312 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005505_050 |
0.0838997481 |
- |
- |
PREDICTED: uncharacterized protein LOC105639309 isoform X2 [Jatropha curcas] |
| 20 |
Hb_106552_050 |
0.0847001792 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |