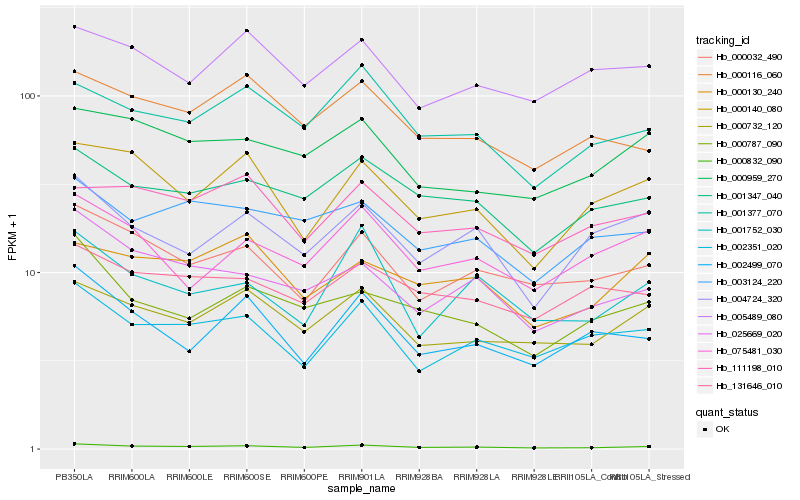
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000832_090 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 2 |
Hb_000732_120 |
0.0698799808 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000032_490 |
0.0725415778 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 4 |
Hb_002351_020 |
0.078326106 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001347_040 |
0.0818896888 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 6 |
Hb_001377_070 |
0.0846320261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000959_270 |
0.0868911705 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 8 |
Hb_000140_080 |
0.0886463341 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_025669_020 |
0.0890191059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000116_060 |
0.0905511693 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 11 |
Hb_003124_220 |
0.0940785494 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 12 |
Hb_002499_070 |
0.0955186027 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004724_320 |
0.09588132 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 14 |
Hb_000130_240 |
0.0972938279 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 15 |
Hb_131646_010 |
0.0973740479 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_005489_080 |
0.0974637741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001752_030 |
0.0978497737 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 18 |
Hb_111198_010 |
0.0981568573 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000787_090 |
0.1001636999 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |
| 20 |
Hb_075481_030 |
0.1001809843 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |