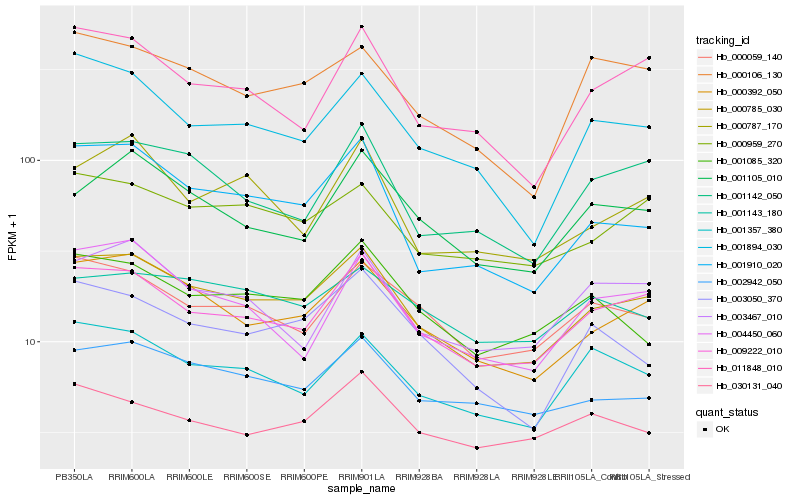
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000785_030 |
0.0 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 2 |
Hb_000392_050 |
0.0529712638 |
- |
- |
PREDICTED: uncharacterized protein LOC105640540 isoform X2 [Jatropha curcas] |
| 3 |
Hb_009222_010 |
0.055139497 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000959_270 |
0.0757076636 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 5 |
Hb_002942_050 |
0.0779743568 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001910_020 |
0.0810034189 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 7 |
Hb_000059_140 |
0.0823554216 |
- |
- |
rnase h, putative [Ricinus communis] |
| 8 |
Hb_001357_380 |
0.0836290943 |
- |
- |
PREDICTED: probable tetraacyldisaccharide 4'-kinase, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_001142_050 |
0.0864156599 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 10 |
Hb_000106_130 |
0.0881585363 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001894_030 |
0.0896055954 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 12 |
Hb_001085_320 |
0.0906870731 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_001105_010 |
0.0910057452 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_011848_010 |
0.0920446028 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 15 |
Hb_003467_010 |
0.0939768964 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_004450_060 |
0.0940311616 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001143_180 |
0.0952388257 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 18 |
Hb_003050_370 |
0.095562625 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 19 |
Hb_030131_040 |
0.0965483265 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 20 |
Hb_000787_170 |
0.0971385274 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |