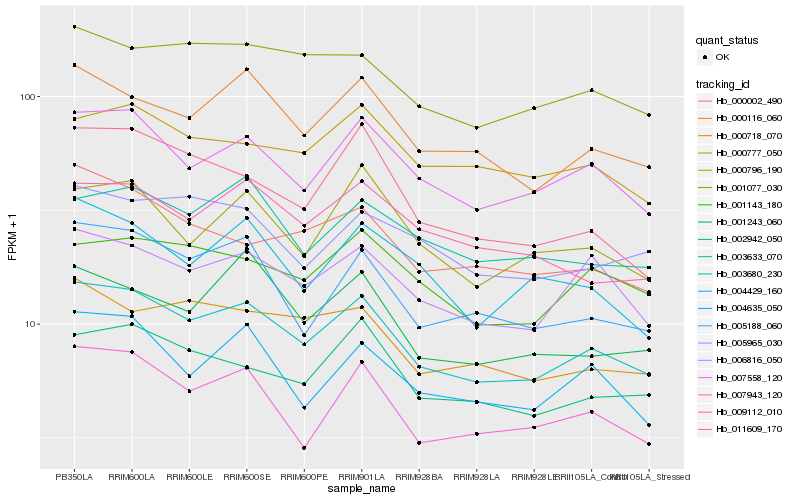
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_230 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 2 |
Hb_007558_120 |
0.0542507896 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 3 |
Hb_000116_060 |
0.0593543485 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 4 |
Hb_006816_050 |
0.0624917582 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 5 |
Hb_002942_050 |
0.0675921672 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 6 |
Hb_005188_060 |
0.0678467363 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 7 |
Hb_007943_120 |
0.068516814 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 8 |
Hb_001077_030 |
0.0715941724 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_004635_050 |
0.0732165003 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 10 |
Hb_000718_070 |
0.0744449023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000796_190 |
0.0754499466 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 12 |
Hb_001143_180 |
0.0756389255 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 13 |
Hb_003633_070 |
0.076957953 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 14 |
Hb_001243_060 |
0.077998453 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 15 |
Hb_004429_160 |
0.0795387027 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000777_050 |
0.0795777167 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 17 |
Hb_005965_030 |
0.0796284942 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 18 |
Hb_000002_490 |
0.0800027068 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 19 |
Hb_009112_010 |
0.080270975 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 20 |
Hb_011609_170 |
0.0806671552 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |