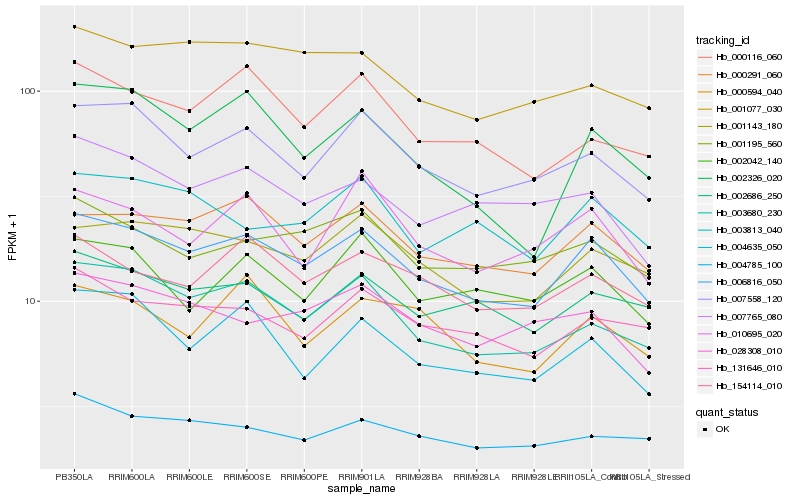
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_050 |
0.0 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X4 [Jatropha curcas] |
| 2 |
Hb_000291_060 |
0.0591157615 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_003680_230 |
0.0624917582 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 4 |
Hb_007558_120 |
0.0626349002 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 5 |
Hb_001143_180 |
0.066837971 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 6 |
Hb_154114_010 |
0.0743190577 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 7 |
Hb_131646_010 |
0.074949915 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002686_250 |
0.0758550202 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_001195_560 |
0.0785340466 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001077_030 |
0.0799793527 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004635_050 |
0.0819245332 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 12 |
Hb_004785_100 |
0.0822026366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000116_060 |
0.0830182022 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 14 |
Hb_003813_040 |
0.0831565246 |
- |
- |
PREDICTED: elongator complex protein 5 [Jatropha curcas] |
| 15 |
Hb_002042_140 |
0.0836077038 |
- |
- |
PREDICTED: uncharacterized protein LOC105647570 [Jatropha curcas] |
| 16 |
Hb_028308_010 |
0.0836899709 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007765_080 |
0.0840870649 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000594_040 |
0.0849371502 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 19 |
Hb_010695_020 |
0.085266663 |
- |
- |
PREDICTED: superkiller viralicidic activity 2-like 2 [Jatropha curcas] |
| 20 |
Hb_002326_020 |
0.0853067254 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |