| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
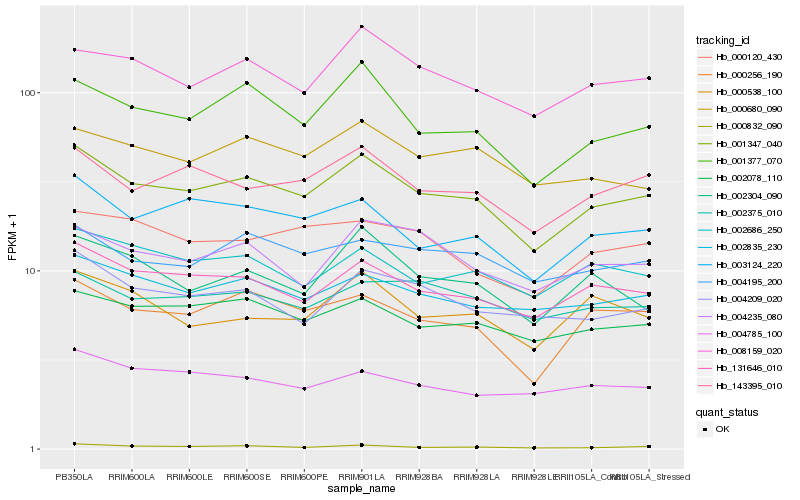
Hb_001347_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 2 |
Hb_131646_010 |
0.0563120675 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_143395_010 |
0.0634171738 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 4 |
Hb_001377_070 |
0.0644283491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003124_220 |
0.0660555547 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 6 |
Hb_000256_190 |
0.0670116677 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002078_110 |
0.0673244898 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 8 |
Hb_008159_020 |
0.0677199909 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 9 |
Hb_002835_230 |
0.0680514737 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_002686_250 |
0.0746010727 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 11 |
Hb_004209_020 |
0.0746678411 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004195_200 |
0.0766489277 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 13 |
Hb_000680_090 |
0.0767392136 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 14 |
Hb_000120_430 |
0.0798186341 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 15 |
Hb_004235_080 |
0.0803427328 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 16 |
Hb_002304_090 |
0.0814998641 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000538_100 |
0.0815272389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004785_100 |
0.081556236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000832_090 |
0.0818896888 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 20 |
Hb_002375_010 |
0.082098487 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |