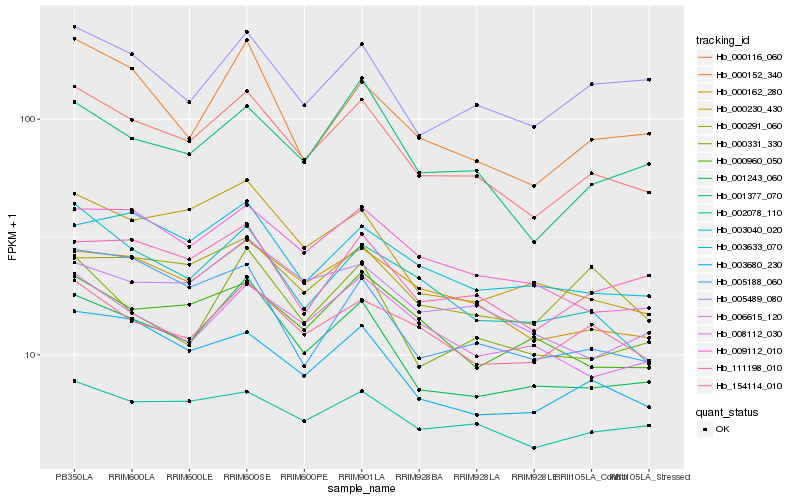
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_060 |
0.0 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 2 |
Hb_001243_060 |
0.0564725153 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 3 |
Hb_003680_230 |
0.0593543485 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 4 |
Hb_000162_280 |
0.0604506942 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 5 |
Hb_001377_070 |
0.0673653013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009112_010 |
0.0709277726 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 7 |
Hb_003040_020 |
0.0714805367 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 8 |
Hb_111198_010 |
0.0754294324 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_154114_010 |
0.0761001572 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 10 |
Hb_003633_070 |
0.0762273577 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 11 |
Hb_000331_330 |
0.076540019 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005489_080 |
0.0789848447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000230_430 |
0.0793097395 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000152_340 |
0.0795442047 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 15 |
Hb_005188_060 |
0.0801078314 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 16 |
Hb_008112_030 |
0.0810117111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002078_110 |
0.0816398136 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 18 |
Hb_006615_120 |
0.0820452436 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 19 |
Hb_000291_060 |
0.082223629 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 20 |
Hb_000960_050 |
0.0824209577 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |