| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
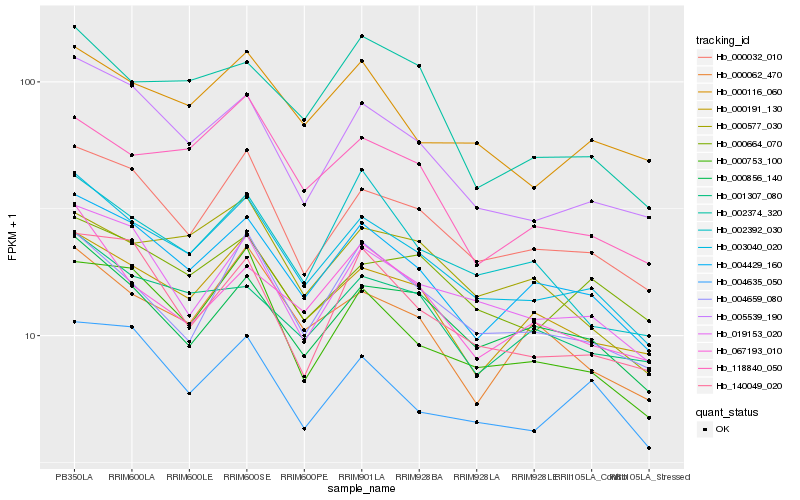
Hb_003040_020 |
0.0 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 2 |
Hb_004429_160 |
0.0497322019 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000032_010 |
0.051794267 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 4 |
Hb_005539_190 |
0.0609525652 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 5 |
Hb_000191_130 |
0.0638696995 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 6 |
Hb_019153_020 |
0.0662426303 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000856_140 |
0.066585521 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004659_080 |
0.0713828032 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 9 |
Hb_000753_100 |
0.0713858984 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640677 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000116_060 |
0.0714805367 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 11 |
Hb_001307_080 |
0.0735432576 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 12 |
Hb_000062_470 |
0.076829704 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 13 |
Hb_067193_010 |
0.0785361632 |
- |
- |
- |
| 14 |
Hb_004635_050 |
0.0800207966 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_118840_050 |
0.0803039969 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 16 |
Hb_000664_070 |
0.0818052418 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 17 |
Hb_140049_020 |
0.0823464205 |
- |
- |
PREDICTED: cell division cycle protein 73 [Jatropha curcas] |
| 18 |
Hb_002392_030 |
0.0829485711 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 19 |
Hb_000577_030 |
0.0831591787 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 20 |
Hb_002374_320 |
0.0834490898 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |